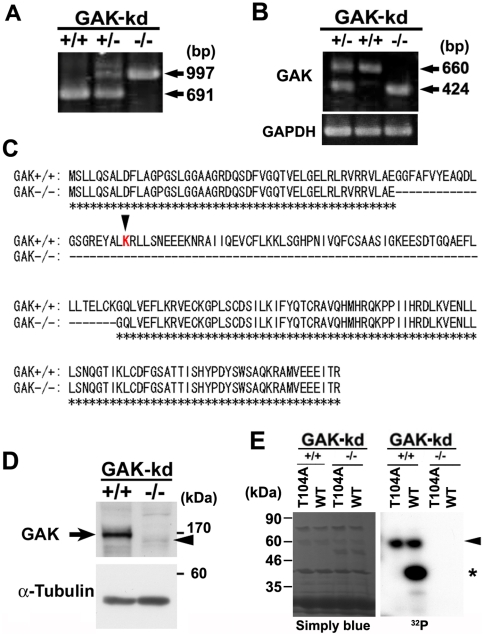
Figure 2. Phenotypes of GAK-/- MEFs.
(A) PCR analysis of genomic DNA of MEFs from heterozygote intercrosses. (B) RT-PCR analysis showing expression of GAK-wt (660 bp) and GAK-kd (424 bp) in GAK+/+, GAK+/-, and GAK-/- MEFs (upper panel). GAPDH was used as a loading control (bottom panel). (C) Amino acid sequence comparisons between prospective translational products from GAK-wt (GAK+/+) and GAK-kd (GAK-/-) genes. Deleted amino acids in GAK-kd (GAK-/-) are indicated by dashed lines. Identical amino acids are indicated by asterisks. A red letter, K, indicates a lysine residue in the ATP-binding site essential for the kinase activity. (D) Western blot analysis of the cell extracts of GAK-wt and -kd MEFs with an anti-GAK polyclonal antibody. An arrow and an arrowhead indicate the GAK-wt and GAK-kd protein, respectively. (E) In vitro kinase assay using purified GST-fused polypeptides corresponding to each kinase domain of GAK derived from GAK-wt and -kd MEFs. The kinase domain of GAK-kd is partially deleted as shown in C. Purified proteins from the PP2A-B'γ subunit (WT and T104A) were used as suitable substrates (indicated with an asterisk). Staining with Coomassie Brilliant Blue G250 (Simply Blue™, Invitrogen) shows a loading control (left panel). An arrowhead indicates auto-phosphorylation of GST-GAK.