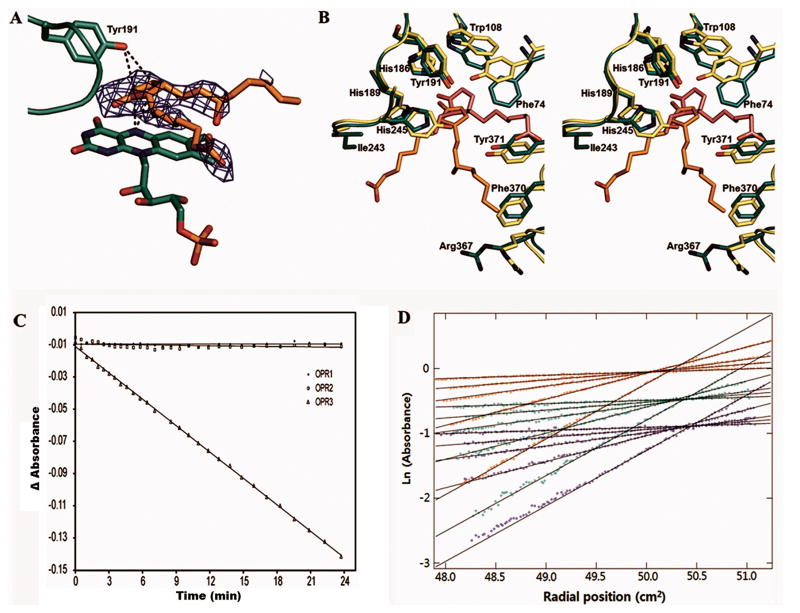
Figure 2.
(A) Orientation of the bound 8-iso-PGA1 relative to the FMN and the general acid Tyr191. The Fo − Fc electron density of the bound 8-iso-PGA1 is contoured at 2.5 σ. The interactions of the double bond-forming carbon atoms of 8-iso-PGA1 with N5 nitrogen atom of FMN and the general acid Tyr191 are indicated by dotted lines.
(B) Stereo view of the substrate binding site of the 8-iso-PGA1-FMN-AtOPR3 complex structure (green) overlaid with the structure of (9R,13R)-OPDA-FMN-LeOPR1 complex (light green) after structural alignment. Residues are numbered according to the AtOPR3 sequence. The FMN molecules of the enzymes superimpose perfectly with each other and are left out for clarity.
(C) Enzymatic assay results of three A. thaliana OPR isoforms using 8-iso-PGA1 as a substrate. Time-dependent change in A340 observed from AtOPR1, AtOPR2, and AtOPR3 are indicated by circles, rectangles, and triangle, respectively.
(D) Sedimentation equilibrium data for three concentrations of AtOPR3 (blue, 4,6 μM; green, 11.5 μM; red, 18.5 μM) and 5 speeds (4.2k, 6.6k, 8.6k, 12k, 17.5k) detected at 278 nm. Global curve fitting was consistent with the presence of a single species at all concentrations observed. The average molecular weight from the global fit of data to the 278 nm data was 44,101 ± 41 Da for AtOPR3. These results are in good agreement with the molecular weight from sequence of 42,694 Da. Data obtained at other wavelengths (378 nm or 461 nm, corresponding to FAD absorption peaks) were consistent with this analysis and their inclusion in global fits minimally impacted the calculated weight average molecular weights.