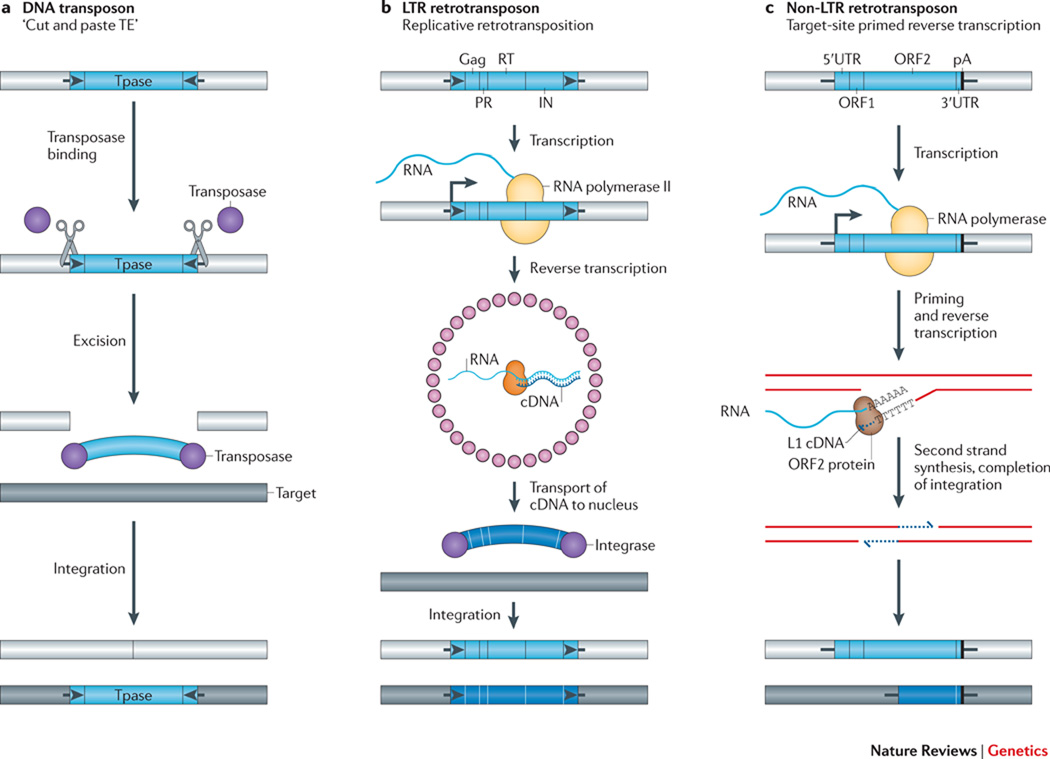
Figure 1. The diverse mechanisms of transposon mobilization.
A. DNA transposons: Many DNA transposons are flanked by terminal inverted repeats (TIRs, black arrows), encode a transposase (TPase, purple circles), and mobilize by a “cut and paste” mechanism (scissors). Transposase binds at or near the TIRs, excises the transposon from its existing genomic location (light gray bar), and pastes it into a new genomic location (dark gray bar). The cleavages of the two strands at the target site are staggered, resulting in a target site duplication (TSD), typically 4 to 8 bp as specified by the TPase (Short black lines flanking the TE). B & C. Retrotransposons: Retrotransposons mobilize by a replicative mechanism that requires the reverse transcription of an RNA intermediate. B. LTR-retrotransposons contain two long terminal repeats (LTRs, black arrows) and encode Gag, protease (PR), reverse transcriptase (RT), and integrase (IN) activities critical for retrotransposition. The 5’ LTR contains a promoter that is recognized by the host RNA polymerase II and produces the mRNA of the TE (start of transcription indicated by a black vertical line attached to a right facing arrow). In step #1 of the reaction, Gag (small pink circles) assembles into virus like particles containing TE mRNA, RT, and IN. The RT copies the TE mRNA into a full-length double stranded DNA (wide blue arc). In step #2 of the reaction, IN (purple circles) inserts the DNA into the new target site. Similar to the TPases of DNA transposons, INs create staggered cuts at the target sites that result in TSDs. C. Non-LTR retrotransposons (right) lack LTRs and encode either one or two open reading frames. The transcription of non-LTR retrotransposons (indicate arrow as in panel b) also leads to the production of a full-length mRNA (blue wavy line). However, these elements mobilize by target-site primed reverse transcription (TPRT). An element-encoded endonuclease generates a single-strand “nick” in genomic DNA, liberating a 3’OH that is used to prime reverse transcription of the RNA. The proteins encoded by autonomous non-LTR retrotransposons also can mobilize non-autonomous retrotransposon RNAs, as well as other cellular RNAs (see text). The TPRT mechanism of an L1 element is depicted in the figure; the new L1 retrotransposition event is 5’ truncated and is retrotransposition-defective (bottom dark gray rectangle with blue line). Some non-LTR retrotransposons lack a poly A tails at their 3’ ends. The integration of non-LTR retrotransposons can lead to target-site duplications (TSDs) and small deletions at the target site in genomic DNA. For example, L1s are generally flanked by 7–20 bp TSDs.