Abstract
Purpose
To investigate the circadian control of the expression of midkine-a and midkine-b in the retina of the zebrafish.
Methods
Zebrafish were maintained in total darkness for 24 hours, and at four-hour intervals, retinas were collected and the expression of mdka during the subjective day and subjective night was evaluated by in situ hybridization, quantitative real-time PCR and Western blot analysis. The circadian expression of mdkb was evaluated by in situ hybridization and real-time PCR.
Results
The expression of mdka increases during the subjective day and decreases during the subjective night. In contrast, the expression of mdkb increases late in the subjective night and decreases late in the subjective day. Within horizontal cells, the two midkine paralogs show asynchronous circadian regulation. Within the annulus of immature retina adjacent to the proliferative margin, mdka shows circadian regulation in Müller glia.
Conclusions
The results of this study show that in the retina of the zebrafish the circadian clock regulates the expression of mdka and mdkb. These growth factors are components of cyclical signaling events with the vertebrate retina. The asynchronous expression of mdka and mdkb in horizontal cells suggests these factors may exert different biological activities at distinct times during the circadian cycle. The circadian control of mdka expression in immature Müller glia may be related to the persistent neurogenesis mediated by these cells.
Indexing terms: mdka, mdkb, horizontal cells, circadian cycle
Introduction
The circadian clock maintains intrinsic rhythmical changes of biochemical and physiological processes, which provide optimal adaptation to environmental changes, such as light, temperature and access to food. In sighted animals, the retina is the primary tissue that entrains the circadian clock to changes in the dark/light cycle [1]. This occurs through direct projections of retinal ganglion cells to the suprachiasmatic nucleus, the master circadian pacemaker, indirect visual projections to the pineal gland [2,3], and through the synthesis of endocrine/paracrine factors, such as melatonin and dopamine [4,5]. Numerous processes in the retina are dependent on circadian rhythms, such as retinomotor movements [6, 7], disc-shedding of photoreceptor outer segments [8], visual sensitivity [9], dopamine release [5], the expression of interphotoreceptor retinoid binding protein [10], and photoreceptor input to cone horizontal cells [11,12,13].
Midkine (MK) is a member of the family of secreted heparin-binding growth/differentiation factors that also includes pleiotrophin [14,15]. MK was first identified in a screen of retinoic acid inducible genes in embryonic carcinoma cells [16,17]; it is highly conserved throughout the animal kingdom and has numerous functions: neurogenic, transforming, neurotrophic, chemotactic, mitogenic, and anti-apoptotic [15,18,19]. In mammals, MK is expressed in numerous tissues during embryonic development, most prominently in the developing neural tube and at epithelial–mesenchymal boundaries [20].
The zebrafish genome encodes two distinct midkine genes, midkine-a (mdka) and midkine-b (mdkb), which during early development have distinct cellular patterns of expression and different biological functions [19,21,22,23]. In the adult retina, mdka is expressed by horizontal cells, whereas mdkb is expressed by ganglion and amacrine cells [24].
Midkines are pleiotropic molecules and little is known about their specific receptors or signaling pathways in zebrafish. To study the specific functions of midkines in the in vivo retinal environment, we began with a thorough characterization of the cellular patterns of expression during various physiological states. Here we report that in horizontal cells quantitative levels of mdka mRNA and protein are regulated by the circadian clock: mdka expression increases during subjective day and decreases during the subjective night, and Mdka protein synthesis follows the same time course. Qualitative evaluation of in situ hybridization indicates that mdkb is also regulated by the circadian clock. The spatial domain of mdkb expression within the inner nuclear layer contracts during the subjective day and expands during the subjective night. Within horizontal cells, the two Midkine paralogs show asynchronous circadian regulation. Though the functional significance of the circadian regulation Midkine expression is yet to be determined, this study expands our knowledge of the cellular expression and circadian control of soluble signaling molecules in the vertebrate central nervous system.
Methods
Animals
Wild-type zebrafish (Danio rerio), mixed strains and strain AB, 4.5 to 7 months old, were purchased from Aquatica Tropicalis (Plant City, Florida) and acclimated for at least 2 weeks in aquaria at 28.5°C and a 14/10 hour light/dark cycle. For experiments described here, fish were maintained in complete darkness for 24hrs and sacrificed at 4-hour intervals, starting either at 12 AM or at 4 AM. These and the following procedures, except where noted (see below), were repeated in three independent experiments. All animal procedures were approved by the University of Michigan Use and Care of Animals in Research Committee.
Tissue preparation for in situ hybridization and immunohistochemistry
At selected times and using dim red illumination, adult fish were anesthetized in 0.1% 3-aminobenzoic acid-ethyl ester (Sigma-Aldrich, St. Louis, MO) until gill movements stopped and sacrificed by cervical transection. Eyes were enucleated, lenses removed and eyecups fixed by immersion for 4–19h at 4°C in 4% paraformaldehyde in 0.1M phosphate buffer containing 5% sucrose. Eyecups were cryoprotected in 20% sucrose, embedded in 2 parts 20% sucrose 1 part Tissue-Tek® O.C.T. Compound (Electron Microscopy Sciences, Hatfield, PA) frozen in Tissue-Tek® OCT and stored at −80°C. Ten-micron thick cryosections through the dorso-ventral axis of the eyecups were mounted on Superfrost Plus microscope slides (Fisher-Scientific, Pittsburgh, PA) and processed for in situ hybridization. Antisense digoxigenin-labeled riboprobes were synthesized using the DIG RNA Labeling kit (Roche Diagnostics, Indianapolis, IN) from plasmids containing the full-length mdka or mdkb cDNAs (gift from Dr. Christoph Winkler [19]). In situ hybridization was performed as described previously [25]. Nitroblue tetrazolium chloride/5-Bromo-4-chloro-3-indolyl phosphate (NBT/BCIP) (Roche Diagnostics, Indianapolis, IN) was used as enzymatic substrate. To compare mdka or mdkb expression across time points, 4–5 sections from eyes collected at each of the six time points were mounted and processed on the same slide. Therefore, within each replicated experiment, sections were processed identically, and this allowed direct comparison of expression levels based on the intensity and spatial distribution of the color reaction.
Photographic images
Images were taken with a Nikon DMX 1200 digital camera mounted on a Nikon Eclipse E800 epifluorescent microscope equipped with a differential interference contrast filter. Adobe Photoshop CS2 (Adobe Systems, San Jose, CA) was used to construct the figures. The layer tool was used to generate overlays. If needed, brightness and contrast were adjusted identically in all panels of a given figure.
RNA extraction and Quantitative Real-Time RT-PCR (QRT-PCR)
To isolate retinal RNA, eyecups were removed and retinas were dissected and carefully separated from the retinal pigment epithelium. Three to four retinas per sample from different zebrafish were pooled for each sample, homogenized with a sterile pestle (Kontes Pellet Pestle, Fisher Scientific) in 200 μl lysis buffer from the Ambion RNAqueous-Micro RNA isolation kit (Ambion, Austin, TX), and RNA extraction was performed according to the manufacturer’s instruction. The amount of RNA was quantified using a spectrophotometer, and the quality of the RNA was assessed on ethydium bromide-stained formalin-agarose gels. 0.5 or 1 μg of total RNA was used to synthesize cDNA using the Superscript II First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) following the manufacturer’s protocol with random hexamers. The resulting first-strand reaction was used as a template for the subsequent QRT-PCR using the iQ™ SYBR® Green Supermix (Bio-Rad, Hercules, CA) in an iCycler Real-Time PCR detection system (Bio-Rad). The following amplification and melt curve analysis protocol was used: 95°C 3min, 35X(95°C: 20s, 57°C: 20s, 72°C: 30s), 95°C: 1min, 90X55°C: 10s. Gene specific primers (0.4μM) were as follows: for mdka (NM_131070) forward: tgaagttttgttactgagctttgtg, and reverse: agccagtgtacataagtgtgtgtgt; for mdkb (NM_131716) forward: gctgttgtaatttgtagcaggtttt, and reverse: cattcaatctcgttgtcatttacag. Serial dilutions of the first strand reaction were run for efficiency calculations of each primer using the Pfaffl method [26]. The threshold cycle (Ct) was determined by the iCycler using the maximum curvature approach and then maintained constant for subsequent runs. Relative gene expression values were determined using the calculated primer efficiencies and threshold cycle with the formula: E−Ct. Specificity of the amplification products was verified by agarose gel electrophoresis of sample wells. Values obtained were averaged for 3–6 independent amplification reactions and statistical significance was calculated by one-way ANOVA with Bonferroni correction for multiple comparisons using SPSS software. Samples within each replicate experiment were treated identically. Results were normalized to the 8 PM time point, which allowed comparison between experiments. To verify that equal amounts of RNA were used, QRTPCR was performed with specific primers for ribosomal protein L-19 (rpl-19, accession number: NM_213208, primers forward: gagtatgctcagacttcagaagagg and reverse: atcaaaccatccttcaccaacttac). There were no differences in rpl-19 expression across time-points.
Midkine-a Antibodies and Western Blot Analysis
Based on antigenicity, hydrophilicity, flexibility and surface probability (Invitrogen, Camarillo, CA), a C-terminal peptide composed of 16 amino acid residues was chosen as the Mdka immunogen (amino acids 131–145: KVKNKPKGKKGKGKGC; accession number NP_571145). Affinity purified polyclonal antibodies were generated in rabbits. In Western blots, the resulting antibodies recognized a single band with the correct size in lysates from both transfected 293T cells expressing zebrafish Mdka-MYC and retinas (data not shown). These antibodies did not recognize Mdkb-MYC (data not shown).
To assay endogenous Mdka, 5 retinas were dissected from 5 different fish at each time-point and pooled. The retinal pigment epithelium was carefully removed, and retinas were homogenized with a Kontes pellet pestle (Fisher Scientific, Pittsburgh, PA) in 75μl of lysis buffer (Phosphate Buffered Saline with 1% Triton X) and protease inhibitors (Complete-mini EDTA free, Roche, Indianapolis, IN). Lysates were centrifuged for 5 minutes at 5000 rpm at 4°C to pellet the nuclei and transferred to fresh tubes. The amount of retinal protein was determined with a BCA protein assay kit (Pierce, Rockford, IL). Equal amounts of protein were loaded onto a 12% SDS-PAGE and processed for immunoblotting.
Proteins were separated by electrophoresis on a 12% Sodium-Dodecyl-Sulphate Polyacrylamide Gel (SDS-PAGE) and transferred onto nitrocellulose membranes (Schleicher and Schuell, Keene, NH). Membranes were incubated at 4°C for at least 4 hours in Blocking Buffer (Phosphate-Buffered-Saline with 0.5% Tween [PBST] and 5% Non-Fat Dry Milk) followed by overnight incubation in αMdka primary antibodies, diluted 1:500. Membranes were washed for one hour in PBST, then incubated for one hour in peroxidase conjugated anti-rabbit IgG secondary antibodies, diluted 1:1000 (Amersham Biosciences, Arlington Heights, IL). Following 4–5 vigorous washes in PBST, proteins were visualized with the electro-chemiluminescence ECL detection system (ECL- Amersham Biosciences, Arlington Heights, IL) and radiographic film (Kodak, Rochester, NY). Antibodies against Mdka recognize a single band at the appropriate molecular weight for the native protein (see Results). Equal protein loading was verified by reprobing membranes with a monoclonal antibody recognizing α– β– and γ-actin (JLA20, Calbiochem, San Diego, CA).
A flatbed scanner (Epson expression 1600) was used to obtain a digital image of the film from which the intensity of the Midkine and actin bands were measured using the histogram tool in Adobe Photoshop CS2 (Adobe Systems, San Jose, CA). The intensity of each Midkine band was normalized to the intensitiy of the corresponding actin band. To compare results between replicate experiments, the 8pm time-point was chosen as a reference, and the relative protein levels were normalized. The resulting values were then averaged between experiments and plotted as fold-change.
Results
The circadian clock regulates expression of mdka and the levels of Mdka protein
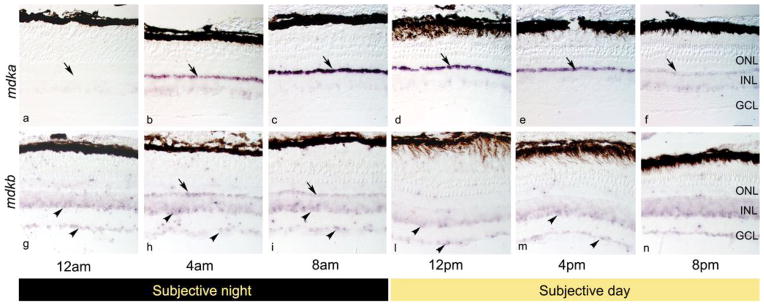
To determine if mdka expression is regulated by the circadian clock, zebrafish were maintained in their normal aquaria, but in total darkness, for a period of 24 hours, and retinas were collected at four-hour intervals. At each time point, retinas were divided into three groups, and the expression of mdka was evaluated by in situ hybridization and QRT-PCR, and the synthesis of Mdka was evaluated by Western blot analysis. The in situ hybridization confirmed that in the adult retina, mdka is selectively expressed in horizontal cells [see 24], and showed that this cellular expression is dynamically modulated during the circadian cycle. Minimum expression was observed in retinas harvested at 12 AM (Fig. 1a), one hour after lights are turned off in our facility. Following this time point, expression increased to reach an apparent maximum at 8 AM (Fig. 1b,c), one hour prior to light onset. mdka expression then remained high during the first half of the subjective day (Fig. 1d,e) and gradually decreased during the second half of the subjective day (Fig. 1f,a), approaching the minimum at midnight.
Figure 1. The circadian clock regulates expression of mdka and mdkb in the zebrafish retina.
Panels a–f are in situ hybridization that illustrate retinal expression of mdka during subjective day (panels a–c) and subjective night (panels d–f). Panels g–n are in situ hybridizations that illustrate retinal expression of mdkb during subjective day (panels g–i) and subjective night (panels l–n). Arrows in panels a–n point to the location of horizontal cells. Arrowheads in panels g–n point to the location of amacrine and ganglion cells respectively. Sections for all time-points for each probe were processed on the same slide. ONL; outer nuclear layer, INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar in f equals 50 μm.
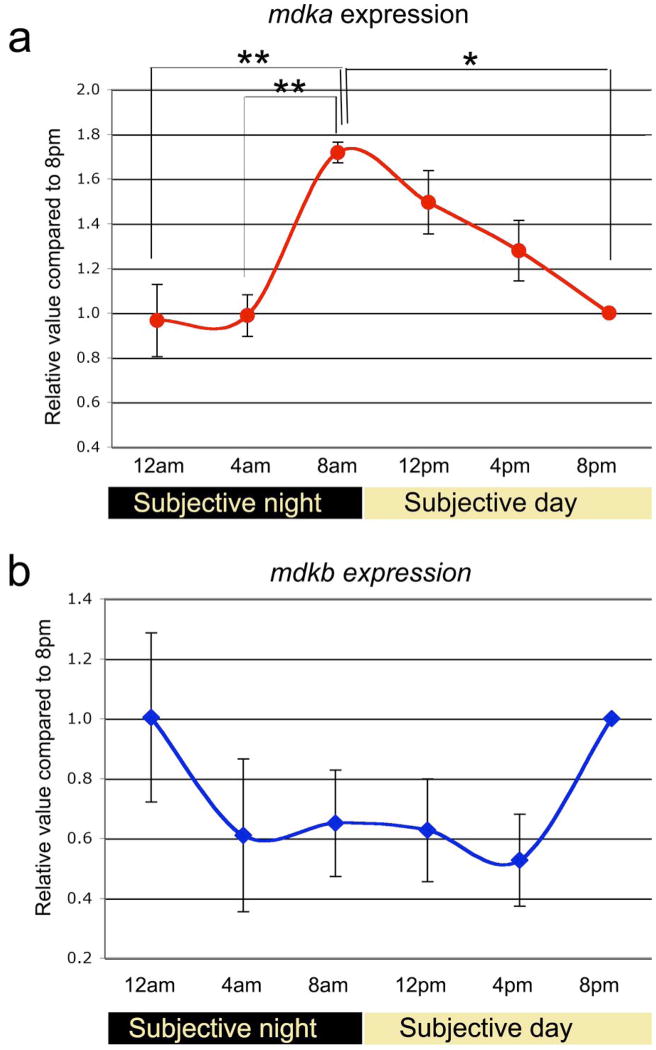
To validate these qualitative observations, QRT-PCR was used to measure mdka mRNA levels. These quantitative data parallel the in situ hybridization data and confirm the circadian rhythm of mdka expression. The ANOVA showed that the expression levels mdka at 12 AM and 4 AM were statistically significantly different from the 8 AM time point, which, in turn, was significantly different from 8 PM (Table 1).
Table 1.
Circadian expression of mdka in the zebrafish retina
| Experiment | 12 am | 4 am | 8 am | 12 pm | 4 pm | 8 pm | Min | Max |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.6599 | 0.8043 | 1.6980 | 1.2189 | 1.1373 | 1 | 0.6599 (12am) | 1.6980 (8am) |
| 2 | 1.0282 | 1.0572 | 1.6504 | 1.6875 | 1.1493 | 1 | 1.0282 (12 am) | 1.6875 (12pm) |
| 3 | 1.2099 | 1.1042 | 1.8055 | 1.5799 | 1.5501 | 1 | 1.1042 (4am) | 1.8055 (8am) |
|
| ||||||||
| Mean | 0.9660 | 0.9886 | 1.7180 | 1.4955 | 1.2789 | 1 | ||
| Std. Dev. | 0.2802 | 0.1613 | 0.0794 | 0.2455 | 0.235 | 0 | ||
| Std. Error | 0.1618 | 0.0931 | 0.0458 | 0.1417 | 0.1356 | 0 | ||
| 95% CI lower bound | 0.2698 | 0.5879 | 1.5207 | 0.8856 | 0.6952 | 1 | ||
| 95% CI upper bound | 1.6623 | 1.3893 | 1.9153 | 2.1054 | 1.8627 | 1 | ||
| One way ANOVA with Bonferroni correction for multiple comparisons
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Pairwise comparison | Mean difference | p value | Pairwise comparison | Mean difference | p value | Pairwise comparison | Mean difference | p value |
| 12am vs 4am | −0.02253 | 1.000 | 8am vs 12am | 0.75197 | 0.007** | 4pm vs 12am | 0.31290 | 1.000 |
| 12am vs 8am | −.75197 | 0.007** | 8am vs 4am | 0.72943 | 0.009** | 4pm vs 4am | 0.29037 | 1.000 |
| 12am vs 12pm | −0.52947 | 0.089 | 8am vs 12pm | 0.2225 | 1.000 | 4pm vs 8am | −0.43907 | 0.255 |
| 12am vs 4pm | −0.3129 | 1.000 | 8am vs 4pm | 0.43907 | 0.255 | 4pm vs 12pm | −0.21657 | 1.000 |
| 12am vs 8pm | −0.03397 | 1.000 | 8am vs 8pm | 0.71800 | 0.01* | 4pm vs 8pm | 0.27893 | 1.000 |
|
| ||||||||
| 4am vs 12am | 0.02253 | 1.000 | 12pm vs 12am | 0.52947 | 0.089 | 8pm vs 12am | 0.03397 | 1.000 |
| 4am vs 8am | −0.72943 | 0.009** | 12pm vs 4am | 0.50693 | 0.115 | 8pm vs 4am | 0.1143 | 1.000 |
| 4am vs 12pm | −0.50693 | 0.115 | 12pm vs 8am | −0.22250 | 1.000 | 8pm vs 8am | −0.718 | 0.01* |
| 4am vs 4pm | −0.29037 | 1.000 | 12pm vs 4pm | 0.21657 | 1.000 | 8pm vs 12pm | −0.4955 | 0.132 |
| 4am vs 8pm | −0.01143 | 1.000 | 12pm vs 8pm | 0.49550 | 0.132 | 8pm vs 4pm | −0.27893 | 1.000 |
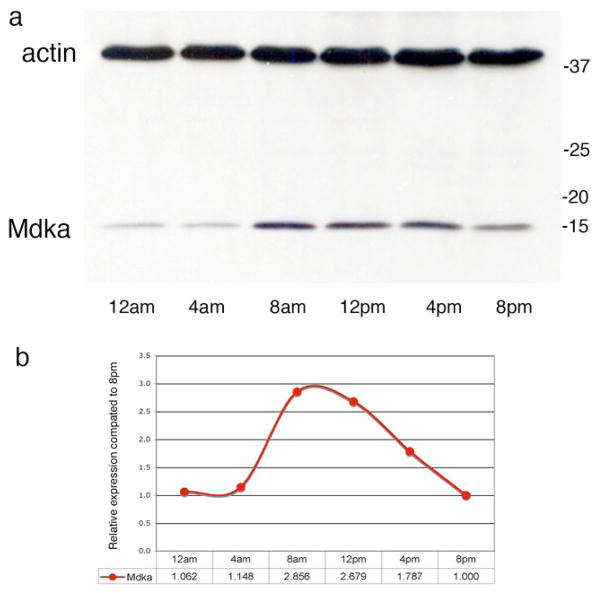
Finally, Western blot analysis of retinal lysates showed a circadian rhythm for protein levels of Mkda that mirror the in situ hybridization and QRT-PCR results (Fig. 3a,b). Minimum levels of protein were present in the retinas at 12 AM, whereas maximum protein levels were present in the retinas at 8 AM. This rhythm was observed in two independent experiments. Taken together, these data show that the expression of Mdka in horizontal cells of the zebrafish retina is regulated by the circadian clock, and both the mRNA and protein levels follow the same dynamic time-related pattern.
Figure 3. Western blot analysis of Mdka expression.
Panel a illustrates an immunoblot of retinal lysates obtained from zebrafish at specified times during the circadian cycle and separated by SDS-PAGE. Lower bands represent the Mdka protein and upper bands represent actin, used as loading control. Panel b illustrates the quantification of circadian changes in retinal Mdka protein expression, normalized to expression of actin, average of 2 experiments. Results are shown as mean fold change compared to the expression at 8pm.
mdka is expressed in presumptive Müller glia at the retinal margin in a circadian pattern
In the developing retina, mdka is transiently expressed in Müller glia [24]. In central retina, these non-neuronal cells strongly express mdka between 3 and 5 days post fertilization (dpf). At 5dpf expression of mdka decreases in Müller cells and increases in horizontal cells, where expression is maintained into adulthood. We consistently observed an inverse relationship between the expression of mdka in horizontal cells and presumptive Müller glia within immature retina adjacent to the neurogenic retinal margin. When mdka expression in horizontal cells was at the minimum (12 AM), its expression increased in cells with large somata located in the inner tier of the inner nuclear layer (Fig. 4). Based on their size and laminar position, we infer these cells are Müller glia. In contrast, when mdka expression in horizontal cells was at the maximum (8 AM), there was no visible expression of mdka in Müller glia.
Figure 4. mdka expression in immature retina.
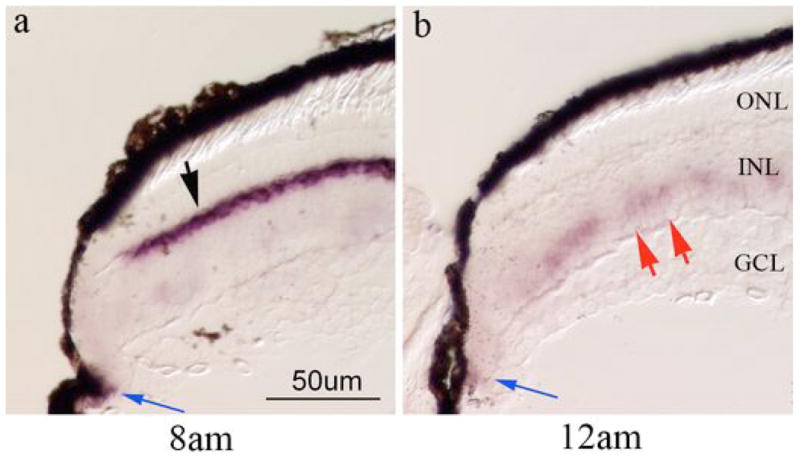
Panels a and b are in situ hybridization that illustrate epression of mdka at the retinal margin at the circadian time of maximum mdka expression (8am, panel a) and minimum mdka expression (12am, panel b). Black arrow in panel a points to horizontal cells that strongly express mdka at this time. Blue arrows in panels a and b point to the location of peripheral-most cells in the CMZ that express mdka. Red arrows in panel b point to the presumptive Müller glia that express mdka at the time when expression of mdka in horizontal cells is minimal. Sections were processed on the same slide. ONL, outer nuclear layer, INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar in f equals 50 μm.
The circadian clock modulates the cellular expression of mdkb
We next asked whether the expression of mdkb is also modulated by the circadian clock. In situ hybridization showed that that at 12 PM mdkb is expressed by cells within the innermost (amacrine cell) tier of the inner nuclear layer and in the ganglion cell layer (Fig. 1j; see also [24]. In contrast, at the onset of the subjective night – 8 PM - expression levels of mdkb increased, and expanded into the outer portion of the inner nuclear layer to include horizontal cells (Fig. 1h,i). This increase in mdkb expression was observed in all three replicate experiments. To quantify mdkb expression QRT-PCR was performed with mdkb-specific primers. In three replicate experiments, a trend that paralleled the in situ hybridizations was observed (Fig. 2b), although the level of mdkb expression was more variable than for mdka and did not reach statistical significance (Table 2).
Figure 2. Quantitative analysis of circadian variations of mdka and mdkb.
Panels a and b are graphical representations of the QRTPCR analysis of the expression of mdka and mdkb, respectively. Gene expression is represented as fold change compared to expression values at the end of the subjective day (8pm). Data represent average values from 3 experiments with standard errors represented for each time-point. Statistical significance was determined through one-way ANOVA with Bonferroni correction for multiple comparisons using SPSS (** p<0.01, * p<0.05).
Table 2.
Circadian expression of mdkb in the zebrafish retina
| Experiment | 12 am | 4 am | 8 am | 12 pm | 4 pm | 8 pm | Min | Max |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.6127 | 0.1035 | 0.2938 | 0.4176 | 0.2846 | 1 | 0.1035 (4 am) | 1 (8pm) |
| 2 | 1.5518 | 0.9176 | 0.834 | 0.9625 | 0.8120 | 1 | 0.8120 (4pm) | 1.55 (12 am) |
| 3 | 0.8471 | 0.8083 | 0.8227 | 0.5001 | 0.4856 | 1 | 0.4846 (4pm) | 1 (8pm) |
|
| ||||||||
| Mean | 1.0039 | 0.6098 | 0.6502 | 0.6242 | 0.5274 | 1 | ||
| Std. Dev. | 048878 | 0.44186 | 0.30867 | 0.29642 | 0.26617 | 0 | ||
| Std. Error | 0.2822 | 0.25511 | 0.17821 | 0.17114 | 0.15368 | 0 | ||
| 95% CI lower bound | −0.2103 | −0.4878 | −0.1166 | −0.1121 | −0.1338 | 1 | ||
| 95% CI upper bound | 2.2181 | 1.7074 | 1.4170 | 1.3605 | 1.1886 | 1 | ||
| One way ANOVA with Bonferroni post-hoc correction for multiple comparisons
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Pairwise comparison | Mean difference | p value | Pairwise comparison | Mean difference | p value | Pairwise comparison | Mean difference | p value |
| 12am vs 4am | 0.39407 | 1.000 | 8am vs 12am | −0.35370 | 1.000 | 4pm vs 12am | − 0.47647 | 1.000 |
| 12am vs 8am | 0.35370 | 1.000 | 8am vs 4am | 0.04037 | 1.000 | 4pm vs 4am | −0.08240 | 1.000 |
| 12am vs 12pm | 0.37967 | 1.000 | 8am vs 12pm | 0.02597 | 1.000 | 4pm vs 8am | −0.12277 | 1.000 |
| 12am vs 4pm | 0.47647 | 1.000 | 8am vs 4pm | 0.12277 | 1.000 | 4pm vs 12pm | −0.968 | 1.000 |
| 12am vs 8pm | 0.00387 | 1.000 | 8am vs 8pm | −0.34983 | 1.000 | 4pm vs 8pm | −0.4726 | 1.000 |
|
| ||||||||
| 4am vs 12am | −0.39407 | 1.000 | 12pm vs 12am | −0.37967 | 1.000 | 8pm vs 12am | −0.00387 | 1.000 |
| 4am vs 8am | −0.4037 | 1.000 | 12pm vs 4am | 0.0144 | 1.000 | 8pm vs 4am | 0.39020 | 1.000 |
| 4am vs 12pm | −0.1440 | 1.000 | 12pm vs 8am | −0.2597 | 1.000 | 8pm vs 8am | 0.34983 | 1.000 |
| 4am vs 4pm | 0.08240 | 1.000 | 12pm vs 4pm | 0.0968 | 1.000 | 8pm vs 12pm | 0.37580 | 1.000 |
| 4am vs 8pm | −0.39020 | 1.000 | 12pm vs 8pm | −0.37580 | 1.000 | 8pm vs 4pm | 0.47260 | 1.000 |
Discussion
The two zebrafish midkine paralogs are encoded by genes located on different chromosomes; they have different cellular patterns of expression in the central nervous system and in the developing hindbrain subserve different functions [19,21,22,23]. In the zebrafish retina, cellular expression of mdka and mdkb is actively modulated during two neurogenic events: retinal development and photoreceptor regeneration following light-induced photoreceptor death [24]. Unexpectedly, a time-course analysis of mdka expression in the light-lesioned retina, consistently showed a marked decrease at twelve hours after light onset. Analyzing expression of mdka in uninjured retinas from zebrafish maintained in normal lighting conditions revealed the same marked decrease in expression at the end of the day (data not shown), suggesting light-induced or circadian regulated expression.
To test whether mdka expression is regulated by the circadian clock we analyzed the expression of mdka and mdkb over a 24hr-period in fish that were kept in total darkness. This analysis confirmed our hypothesis that in the zebrafish retina expression of midkines is regulated by the circadian clock. For mdka, the circadian changes in mRNA expression were paralleled by changes in the levels of Mdka protein, suggesting that mRNA and protein synthesis and degradation are tightly regulated. For mdkb, in situ hybridization showed that mRNA levels for cells in the inner nuclear layer appear also to be modulated by the circadian clock. The QRT-PCR analysis for mdkb expression revealed a trend that mirrored the in situ hybridization, but differences were not statistically significant. Unlike mdka, which is very distinctly expressed in horizontal cells, mdkb is expressed in several types of retinal neurons, so it is possible that the QRT-PCR analysis of whole retina RNA was not able to reflect the circadian changes in expression of mdkb in horizontal cells. Nevertheless, in situ hybridization showed that in horizontal cells, the expression mdka and mdkb is out-of-phase, suggesting that in the outer retina these two proteins exert their biological actions at different times during the circadian cycle.
Interestingly, we also observed the circadian-regulated expression of mdka in Müller glia, but only in a few immature cells found within the growth zone adjacent to the neurogenic retinal margin (see [27] for a more complete description of neurogenic events in the adult retina of teleosts). This observation is reminiscent of the transient expression of mdka in Müller glia in the larval retina and the re-expression of mdka in Müller glia during photoreceptor regeneration [24]. Together, these results suggest that expression of mdka in Müller glia may be related to the persistent (and injury-induced) neurogenesis mediated by these cells [28].
As for most vertebrates, in teleosts the retina functions in part (though not alone, [29,30,31]) to entrain circadian rhythms. Additionally, within the teleost retina numerous processes are under direct circadian regulation. Among these is the synthesis and release of neurotransmitters and soluble signaling molecules [5,32], retinomotor movements [6,7], and the synthesis of photoreceptor opsins [33,34]. Whereas the function of retinal Midkines are not yet known, our data identify them as components of cyclical signaling events with the vertebrate retina. Further, the demonstration that these molecules are regulated by circadian rhythms points to important avenues of further inquiry.
Acknowledgments
We thank Dr. Christoph Winkler for the gift of plasmids encoding mdka and mdkb. We also thank Linda Barthel, Dilip Pawar and Laura Kakuk-Atkins for excellent technical assistance.
This work was supported by NIH grants R01-EY07060 and P30-EY07003 and a Senior Scientific Investigator Award from the Research to Prevent Blindness (PFH)
References
- 1.Nelson RJ, Zucker I. Absence of extraocular photoreception in diurnal and nocturnal rodents exposed to direct sunlight. Comp Biochem Physiol. 1981;69A:145–148. [Google Scholar]
- 2.Moore RY, Speh JC, Card JP. The retinohypothalamic tract originates from a distinct subset of retinal ganglion cells. J Comp Neurol. 1995;352:351–66. doi: 10.1002/cne.903520304. [DOI] [PubMed] [Google Scholar]
- 3.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–41. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 4.Wiechmann AF, Summers JA. Circadian rhythms in the eye: the physiological significance of melatonin receptors in ocular tissues. Prog Retin Eye Res. 2008;27:137–60. doi: 10.1016/j.preteyeres.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Ribelayga C, Wang Y, Mangel SC. A circadian clock in the fish retina regulates dopamine release via activation of melatonin receptors. J Physiol. 2004;554:467–82. doi: 10.1113/jphysiol.2003.053710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pierce ME, Besharse JC. Circadian regulation of retinomotor movements. Interaction of melatonin and dopamine in the control of cone length. J Gen Physiol. 1985;86:671–89. doi: 10.1085/jgp.86.5.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menger GJ, Koke JR, Cahill GM. Diurnal and circadian retinomotor movements in zebrafish. Vis Neurosci. 2005;22:203–9. doi: 10.1017/S0952523805222083. [DOI] [PubMed] [Google Scholar]
- 8.LaVail MM. Circadian nature of rod outer segment disc shedding in the rat. Invest Ophthalmol Vis Sci. 1980;19:407–11. [PubMed] [Google Scholar]
- 9.Li L, Dowling JE. Zebrafish visual sensitivity is regulated by a circadian clock. Vis Neurosci. 1998;15:851–7. doi: 10.1017/s0952523898155050. [DOI] [PubMed] [Google Scholar]
- 10.Rajendran RR, Van Niel EE, Stenkamp DL, Cunningham LL, Raymond PA, Gonzalez-Fernandez F. Zebrafish interphotoreceptor retinoid-binding protein: differential circadian expression among cone subtypes. J Exp Biol. 1996;199:2775–87. doi: 10.1242/jeb.199.12.2775. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Mangel SC. A circadian clock regulates rod and cone input to fish retinal cone horizontal cells. Proc Natl Acad Sci. 1996;93:4655–60. doi: 10.1073/pnas.93.10.4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ribelayga C, Wang Y, Mangel SC. Dopamine mediates circadian clock regulation of rod and cone input to fish retinal horizontal cells. J Physiol. 2002;544:801–16. doi: 10.1113/jphysiol.2002.023671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ribelayga C, Cao Y, Mangel SC. The circadian clock in the retina controls rod-cone coupling. Neuron. 2008;59:790–801. doi: 10.1016/j.neuron.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obama H, Matsubara S, Guénet JL, Muramatsu T. The midkine (MK) family of growth/differentiation factors: structure of an MK-related sequence in a pseudogene and evolutionary relationships among members of the MK family. J Biochem. 1994;115:516–22. doi: 10.1093/oxfordjournals.jbchem.a124368. [DOI] [PubMed] [Google Scholar]
- 15.Kadomatsu K, Muramatsu T. Midkine and pleiotrophin in neural development and cancer. Cancer Lett. 2004;204:127–43. doi: 10.1016/S0304-3835(03)00450-6. [DOI] [PubMed] [Google Scholar]
- 16.Kadomatsu K, Tomomura M, Muramatsu T. cDNA cloning and sequencing of a new gene intensely expressed in early differentiation stages of embryonal carcinoma cells and in mid-gestation period of mouse embryogenesis. Biochem Biophys Res Commun. 1988;151:1312–8. doi: 10.1016/s0006-291x(88)80505-9. [DOI] [PubMed] [Google Scholar]
- 17.Kadomatsu K, Huang RP, Suganuma T, Murata F, Muramatsu T. A retinoic acid responsive gene MK found in the teratocarcinoma system is expressed in spatially and temporally controlled manner during mouse embryogenesis. J Cell Biol. 1990;110:607–16. doi: 10.1083/jcb.110.3.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muramatsu T. Midkine and pleiotrophin: two related proteins involved in development, survival, inflammation and tumorigenesis. J Biochem. 2002;132:359–71. doi: 10.1093/oxfordjournals.jbchem.a003231. [DOI] [PubMed] [Google Scholar]
- 19.Winkler C, Schafer M, Duschl J, Schartl M, Volff JN. Functional divergence of two zebrafish midkine growth factors following fish-specific gene duplication. Genome Res. 2003;13:1067–81. doi: 10.1101/gr.1097503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitsiadis TA, Salmivirta M, Muramatsu T, Muramatsu H, Rauvala H, Lehtonen E, Jalkanen M, Thesleff I. Expression of the heparin-binding cytokines, midkine (MK) and HB-GAM (pleiotrophin) is associated with epithelial-mesenchymal interactions during fetal development and organogenesis. Development. 1995;121:37–51. doi: 10.1242/dev.121.1.37. [DOI] [PubMed] [Google Scholar]
- 21.Winkler C, Moon RT. Zebrafish mdk2, a novel secreted midkine, participates in posterior neurogenesis. Dev Biol. 2001;229:102–18. doi: 10.1006/dbio.2000.9967. [DOI] [PubMed] [Google Scholar]
- 22.Schäfer M, Rembold M, Wittbrodt J, Schartl M, Winkler C. Medial floor plate formation in zebrafish consists of two phases and requires trunk-derived Midkine-a. Genes Dev. 2005;19:897–902. doi: 10.1101/gad.336305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liedtke D, Winkler C. Midkine-b regulates cell specification at the neural plate border in zebrafish. Dev Dyn. 2008;237:62–74. doi: 10.1002/dvdy.21384. [DOI] [PubMed] [Google Scholar]
- 24.Calinescu AA, Vihtelic TS, Hyde DR, Hitchcock PF. The Cellular Expression of Midkine-a and Midkine-b During Retinal Development and Photoreceptor Regeneration in Zebrafish. J Comp Neurol. 2009 doi: 10.1002/cne.21999. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hitchcock PF, Kakuk-Atkins L. The basic helix-loop-helix transcription factor neuroD is expressed in the rod lineage of the teleost retina. J Comp Neurol. 2004;477:108–17. doi: 10.1002/cne.20244. [DOI] [PubMed] [Google Scholar]
- 26.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raymond PA, Barthel LK, Bernardos RL, Perkowski JJ. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev Biol. 2006;6:36. doi: 10.1186/1471-213X-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bernardos RL, Barthel LK, Meyers JR, Raymond PA. Late-stage neuronal progenitors in the retina are radial Muller glia that function as retinal stem cells. J Neurosci. 2007;27:7028–40. doi: 10.1523/JNEUROSCI.1624-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vuilleumier R, Besseau L, Boeuf G, Piparelli A, Gothilf Y, Gehring WG, Klein DC, Falcón J. Starting the zebrafish pineal circadian clock with a single photic transition. Endocrinology. 2006;147:2273–9. doi: 10.1210/en.2005-1565. [DOI] [PubMed] [Google Scholar]
- 30.Vallone D, Lahiri K, Dickmeis T, Foulkes NS. Zebrafish cell clocks feel the heat and see the light. Zebrafish. 2005;2:171–87. doi: 10.1089/zeb.2005.2.171. [DOI] [PubMed] [Google Scholar]
- 31.Dekens MP, Whitmore D. Autonomous onset of the circadian clock in the zebrafish embryo. EMBO. 2008;27:2757–65. doi: 10.1038/emboj.2008.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vuilleumier R, Boeuf G, Fuentes M, Gehring WJ, Falcón J. Cloning and early expression pattern of two melatonin biosynthesis enzymes in the turbot (Scophthalmus maximus) Eur J Neurosci. 2007;25:3047–57. doi: 10.1111/j.1460-9568.2007.05578.x. [DOI] [PubMed] [Google Scholar]
- 33.Korenbrot JI, Fernald RD. Circadian rhythm and light regulate opsin mRNA in rod photoreceptors. Nature. 1989;337:454–7. doi: 10.1038/337454a0. [DOI] [PubMed] [Google Scholar]
- 34.Li P, Chaurasia SS, Gao Y, Carr AL, Iuvone PM, Li L. CLOCK is required for maintaining the circadian rhythms of Opsin mRNA expression in photoreceptor cells. J Biol Chem. 2008;283:31673–8. doi: 10.1074/jbc.M803875200. [DOI] [PMC free article] [PubMed] [Google Scholar]