Abstract
DNA molecules provide what is probably the most iconic example of self-replication—the ability of a system to replicate, or make copies of, itself. In living cells the process is mediated by enzymes and occurs autonomously, with the number of replicas increasing exponentially over time without the need for external manipulation. Self-replication has also been implemented with synthetic systems, including RNA enzymes designed to undergo self-sustained exponential amplification1-5. An exciting next step would be to use self-replication in materials fabrication, which requires robust and general systems capable of copying and amplifying functional materials or structures. Here we report a first development in this direction, using DNA tile motifs that can recognize and bind complementary tiles in a pre-programmed fashion. We first design tile motifs so they form a seven-tile seed sequence, then use the seeds to instruct the formation of a first generation of complementary seven-tile daughter sequences, and finally use the daughters to instruct the formation of seven-tile granddaughter sequences that are identical to the initial seed sequences. Considering that DNA is a functional material that can organize itself and other molecules into useful structures6-13, our findings raise the tantalizing prospect that we may one day be able to realize self-replicating materials with various patterns or useful functions.
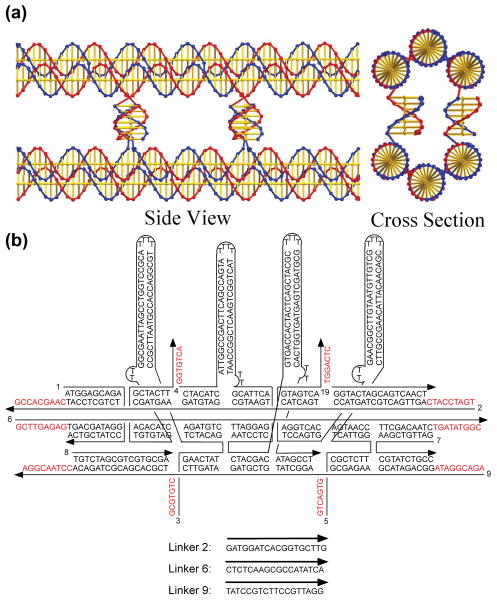
Nucleic acids comprise the genetic material of all living organisms, with the ordered pattern of bases in the DNA double helix readily copied by an enzymatic process that leads to semi-conservative replication within the cell14,15. The replication process exploits the complementarity of the four-letter code made of the bases within the linear DNA double helix, where adenine (A) pairs with thymine (T) and guanine (G) pairs with cytosine (C).16 We transfer this concept to DNA materials, and use as analog of the nucleotide letter the bent triple crossover motif (BTX)17 that displays four single strands (of 7 nucleotides each) to connect with a similar second BTX molecule. Information is encoded using these four strands, which ensure a specific interaction between two BTX molecules and therefore the transfer of information from a seed structure to later generations. As shown in Figure 1a, the structure formed by two paired BTX letters is a paired 6-helix bundle motif (P6HB). It contains six helices connected to each other by four small double helices that extend from one duplex to another.
Figure 1. DNA tile sequences and structures.
(a) The P6HB Motif. This representation24 shows the way in which two BTX domains are paired by four lateral connections to form the P6HB motif. The cross section view shows two of the four helices that are formed by the lateral cohesive interactions. The interactions at the rear are eclipsed in this projection. (b) The Sequence and Structure of the B′ BTX Tile. Four helical domains, hairpins, are shown attached perpendicular to the BTX motif, so that they will create a topographic feature that can be detected in the atomic force microscope (AFM). Other tiles are shown in the Supplementary Information (S1).
Once specific recognition has been set and BTX motifs have been paired on an initial seed as desired, they must be bound together linearly, analogous to the formation of a DNA backbone. This is achieved using 18 nucleotides per duplex (9 from each side of the three BTX double helical domains) and binding them with a solution-derived DNA strand. We use the same DNA sequence in every case (except in the seed), so that any pattern can be replicated. The cohesive strength of the longitudinal bonds linking our tiles to form longer arrays is necessarily of higher melting temperature than the lateral binding between seed and daughter arrays, both because of greater length and because of end-stacking interactions18; this is analogous to the bases in the double helix being strongly attached (covalently bonded) in the direction parallel to the helix axis and weakly attached (hydrogen-bonded) perpendicular to it. The difference in bonding interactions allows a complementary sequence to be assembled on a seed template, internally fused, and then separated from the template to form an autonomous copy. We note that in contrast to the DNA code as we know it, the BTX-based code is not limited to four letters: in principle we can design as many as (428) different combinations of strands4 involved in lateral pairing.
Experimentally, we start with the simplest case of using two different logical BTX species, an A tile, a B tile and their complementary A′, and B′ tiles. Tiles are labeled for AFM identification with either biotinylated nucleotides that bind streptavidin or with large hairpin features (see Fig 1b for a labeled B′ tile). We can thus read with AFM any sequence created from A and B components, and check the fidelity of our replication from the initial seed to the new generations of daughters and granddaughters. The labeling also allows us to verify whether the initial seed patterns has formed accurately. The first tile of the seed is an initiator I (an A-like tile attached to a magnetic bead), followed by a series of A and B tiles to give the sequence IBBABAB. In the seed it is the A tiles that contain labels, while in the complements it is the B′ tiles. In terms of labeling and using L and N to represent the presence and absence of labels, respectively, the seed array should have the sequence LNNLNLN and the newly replicated sequence should appear as NLLNLNL. (Note, though, that any array can be labeled uniquely in any generation.) In addition to creating the seed and its replica, we have also replicated the replica and thereby obtained a direct copy of the information in the seed array itself.
The starting point for each of the stages in the replication process is the self-assembly of BTX tiles. (See supplementary information section S1 for the sequences of every tile used, section S2 for non-denaturing gels that demonstrate that the BTX tiles form properly and only associate as complementary pairs, and section S3 for electrophoretic mobility (Ferguson) analyses of the individual tiles and their P6HB complexes.)
A schematic of the seed assembly is given in Figure 2a. In step 1, the seven different seed tiles are formed separately, by annealing their constituent strands. In step 2, the tiles are mixed so they can assemble into seven-tile seed arrays, with every tile placed in the proper order as a result of specific inter-tile interactions provided by the unique 16-mer sticky ends on each of its helices. In the figure, the sticky ends are labeled to indicate position within the array and binding partner; for example, Y1 on the A1 (I) tile bonds to Y1′ on the B2 tile, leading to an A tile at the first position of the seed and a B tile at the second position. There is a single sticky end on the left of the A1 tile, labeled S, which is used in a later step to bind to a magnetic dynabead. The A tiles contain accessible biotin groups, and in step 3 are labeled with streptavidin for AFM identification. The images in Fig 2b illustrates the successful formation of the seeds: a number of incorrectly formed, multimeric or aggregated complexes are visible, but the modal images are straight linear arrays containing bumpy features, as we expect. The three higher-resolution images in Fig 2b show that the target arrays have been assembled. Note that for replication, we use unpurified seeds that have not been streptavidin labeled.
Figure 2. DNA seeds.
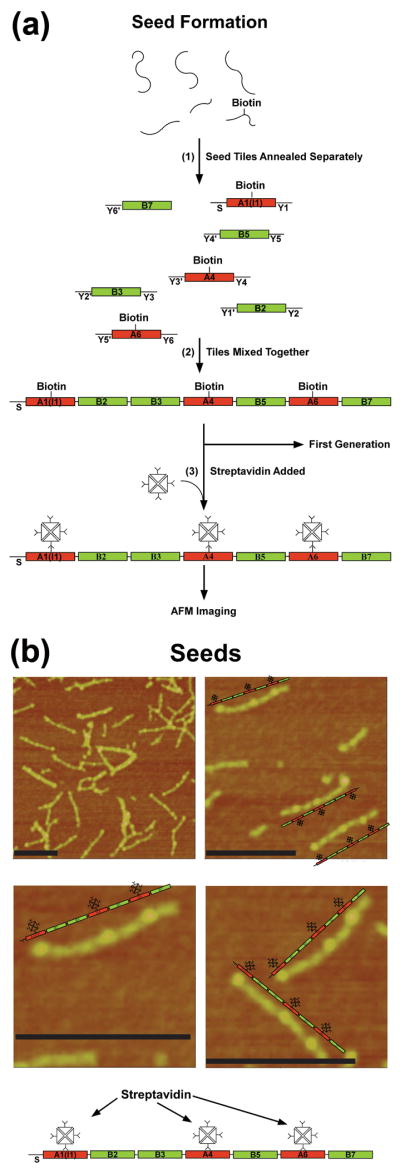
(a) Seed Formation. This drawing shows in step 1 how the individual strands of the seed tiles are self-assembled in separate vessels to produce seven different BTX tiles flanked by three sets of unique sticky ends labeled Y and a number; primed numbers are complementary to unprimed numbers. The red tiles are the A tiles and the green tiles are the B tiles. The A tiles contain a biotin group to enable their decoration by streptavidin. The tile labeled A1(I1) is the initiator tile. The strand labeled S on its left can bind to a dynabead during the replication process. Step 2 shows that these tiles produce 7-unit seeds when they are mixed together. The tiles are prepared for AFM imaging by the addition of streptavidin (Step 3) (b) AFM Images of Seeds. The image at the upper left shows a typical field slightly less than a square micron. Black scale bars correspond to 200nm. A large number of seeds are present, along with some multimeric complexes. The other three panels are zoomed images. A schematic image of each seed is shown next to the seed.
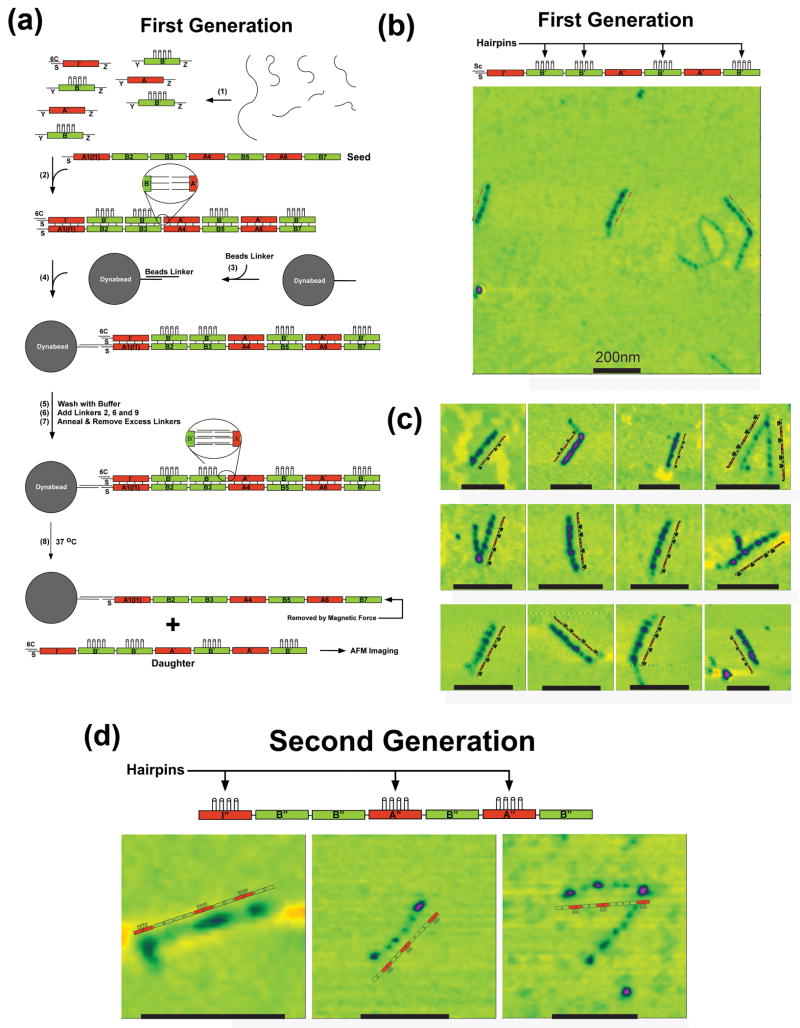
The generation of complementary daughter arrays, schematically outlined in Fig 3a, starts similar to the generation of seed arrays with the self-assembly of A′ and B′ motifs, annealed in different vessels (step 1) and then added to a solution of seed arrays and mixed (step 2). The initiator tile of the daughter generation contains the same S end that was on the initiator tile of the seed, but this strand is protected by a cover strand19 to keep it free of any beads. Otherwise, the tiles are all flanked by the same set of sticky end connectors (nonamers denoted Y and Z in the figure). The B′ tiles contain a group of covalently attached hairpins as labels.
Figure 3. DNA generations.
(a) Replication of the Seed Pattern in the First Generation. Strands are annealed in step 1, where the tiles are all flanked by the same connectors, designated Y and Z. The initiator tile contains a protected S-strand, paired with a cover strand, 6C. The B′ tiles contain the 4-hairpin markers for AFM imaging. In the presence of the seed tile (step 2), the strands assemble into a pattern mimicking the seed pattern. The magnetic dynabead is prepared in step 3, and attached to the seed (step 4). This is followed by a wash step, the addition of linkers and their annealing (steps 5-7). Heating the system to 37 °C results in the separation of the daughter 7-tile complex and the seed (removed magnetically). (b) Atomic Force Microscopy of First Generation Constructs Showing a Typical Field Slightly Larger than a Square Micron. Black scale bars correspond to 200nm. (c) Zoomed Images of Heptameric Daughter Complexes. These images, flanked by explanatory schematic images demonstrate that the ABBABAB pattern has been replicated successfully. (d) AFM Images of Second Generation Molecules. These zoomed images show the pattern that was programmed in the original seed tile.
Following mixing, the BTX tiles with complementary lateral cohesive ends are paired together to form an extended P6HB motif; the result is a complementary daughter array attached to the seed array, with the daughter tiles not yet bound to each other. At this point, a dynabead is introduced containing a linker complementary to the S strand on the seed, so that the entire construct is attached to the bead. Following a wash, 18-mer linkers that are complementary to both of the nonamer sticky ends extending between tiles are added to the solution. These 18-mers are annealed to the construct, so that the connections between the tiles are formed. This attachment is seen in the blown-up part of the system following step 7. Following a wash to remove excess linkers, seeds and daughters are separated by heating the system to 37 °C, where lateral pairs are broken but the longitudinal bonds remain intact (step 8 at bottom of Figure 3a).
Despite the large number of manipulations required to prepare the daughters, the large-field AFM image of this system (Fig 3b) still contains multiple copies of the molecules in a field a little over a square micron, along with some flawed products. Comparison with the image in Fig 2b reveals that the daughters are significantly sparser than the seeds. The zoomed images in Fig 3c show daughter tile arrays with four clear features that thus appear to have formed correctly, although some of them are parts of aggregates. The percentage of heptamers with the correct sequence is estimated to be 70% (23 molecules sampled).
Granddaughters (replicas of the seed arrays) were produced directly from an initial preparation, rather than from purified seeds or purified daughters (see Supplementary Information S4). Cover strands were removed from the initiator tile, and A″ and B″ tiles are added to the solution. In this second generation, A″ tiles contain the hairpin labels. The same sequence of steps is then performed as in the generation of the daughter strands (see Methods and Supplementary Information S4), with daughters and granddaughters separated by heating to 37 °C.
Figure 3d shows AFM images of granddaughters, of which there are fewer molecules and fewer cases of unambiguously correct images than seen with daughters. We estimate the percentage of second-generation heptamers with a correct tile sequence to be 31% (18 molecules sampled), corresponding to 55% (∼0.311/2) per generation, similar to the 70% seen for the daughters (See S5). We note that a net increase of molecules and hence a growing system would be obtained if we were to retain the original seeds in the solution containing the granddaughter molecules.
At present, our replication method is still rather cumbersome as it requires multiple chemical and thermal processing cycles; and unlike the elegant work of Lincoln and Joyce with RNA enzymes1, it does not yet achieve exponential amplification. However, we have demonstrated that it is possible to replicate not just molecules like DNA or RNA, but discrete tertiary structures that could in principle assume many different shapes and functional features. We also note that in the same way that the first cars, airplanes and computers were clunky relative to current systems, it should be possible to make the present procedure smoother and more sophisticated. For example, yield is obviously affected by the removal of seed molecules to produce the daughters and likewise by the removal of daughters to produce the granddaughters so as to simplify analysis. This could be avoided through elimination of bead removal steps, by using self-protected hairpins20 or photoactive molecules for the longitudinal interactions. We expect that this and other improvements will deliver a robust replication method that is applicable to molecular, nanometer-sized and colloidal systems (such as patchy21 and lock and key22 particles) displaying programmed recognition.
Methods Summary
All strands were designed using the program SEQUIN.23 Following PAGE purification, strands for the seeds, daughter and granddaughter tiles were mixed stoichiometrically as estimated by OD260 and dissolved to 0.5 μM in TAE/Mg2+ buffer (40 mM Tris-HCl, 20 mM Acetic Acid, 2 mM EDTA, 12.5 mM Magnesium Acetate, pH 8.0). The solutions were slowly annealed from 90 °C to room temperature (RT) over 48 hours in a 2-litre water bath insulated in a Styrofoam box. Stoichiometric quantities of seven seed tiles were mixed and annealed from 45 °C to RT over 24 hours to make seeds. To form the first generation, three first-generation tiles (I′, A′, and B′) were mixed with annealed first-generation tiles (seeds:I′:A′:B′=1:2:4:8), and slowly annealed from 45 °C to RT. Dynabeads were washed with ddH2O and TAE/Mg buffer, mixed with beads linker in TAE/Mg buffer, slowly annealed from 55 °C to RT, washed with buffer, and mixed with DNA solution. The solution containing dynabeads was annealed from 33 °C to 23 °, placed on a magnetic stand and washed with TAE/Mg buffer. Linking strands 2, 6 and 9 were then added, the solution cooled from 33 °C to 23 °C, placed on a magnetic stand and washed with TAE/Mg buffer to remove excess linkers. Dynabeads in TAE/Mg buffer were kept at 37 °C for one hour, placed on the magnetic stand, and the solution was removed from dynabeads and stored in a clean tube for AFM imaging. Formation of the second generation is similar to the first: It starts from initial seed preparation, followed by formation of the first generation, and adding second-generation tiles (I″, A″, and B″). Steps (2)-(8) described in formation of the first-generation were repeated.
Supplementary Material
Acknowledgments
This research has been partially supported by a grant from the W.M. Keck Foundation, by the MRSEC Program of the National Science Foundation under Award Number DMR-0820341 and NASA NNX08AK04G to PMC, as well as by the following grants to NCS: GM-29554 from the National Institute of General Medical Sciences, CTS-0608889 and CCF-0726378 from the National Science Foundation, 48681-EL and W911NF-07-1-0439 from the Army Research Office, and N000140910181 and N000140911118 from the Office of Naval Research. M.E.L. acknowledges a Netherlands Organization for Scientific Research (NWO) for a Rubicon grant, and C.M. acknowledges a DAAD post-doctoral grant.
Footnotes
Supplementary Information: Sequences of the tiles used; non-denaturing gels of tiles; Ferguson analyses of tiles and their complexes; protocol for producing granddaughter molecules, statistics for producing seeds, daughters and granddaughters.
Author Contributions: T.W. designed experiments, performed experiments, analyzed data and wrote the paper; R.S. designed experiments, performed experiments, analyzed data and wrote the paper; R.D. analyzed data and wrote the paper; M.E.L. initiated the project, analyzed data and wrote the paper; C.M. designed experiments, performed experiments, analyzed data and wrote the paper; D.J.P. initiated and directed the project, analyzed data and wrote the paper; P.M.C. initiated and directed the project, analyzed data and wrote the paper; N.C.S. initiated and directed the project, designed experiments, analyzed data and wrote the paper.
The authors declare no competing financial interests.
Contributor Information
Tong Wang, Email: tongwang@nyu.edu.
Ruojie Sha, Email: ruojie.sha@nyu.edu.
Rémi Dreyfus, Email: dreyfus@nyu.edu.
Mirjam E. Leunissen, Email: m.e.leunissen@amolf.nl.
Corinna Maass, Email: cm162@nyu.edu.
David J. Pine, Email: pine@nyu.edu.
References
- 1.Lincoln TA, Joyce GF. Self-sustained replication of an RNA enzyme. Science. 2009;323:1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wintner EA, Conn MM, Rebek J., Jr Studies in molecular replication. Accts Chem Res. 1994;27:198–203. [Google Scholar]
- 3.Schulman R, Winfree E. Synthesis of crystals with a programmable kinetic barrier to nucleation. Proc Nat Acad Sci (USA) 2007;104:15236–15241. doi: 10.1073/pnas.0701467104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin C, et al. In vivo cloning of artificial DNA nanostructures. Proc Nat Acad Sci (USA) 2008;105:17626–17631. doi: 10.1073/pnas.0805416105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee DH, Severin K, Yokobayashi Y, Ghadiri MR. Emergence of symbiosis in peptide self-replication through a peptide hypercyclic network. Nature. 1997;390:591–594. doi: 10.1038/37569. [DOI] [PubMed] [Google Scholar]
- 6.Seeman NC. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 7.Zhu L, Lukeman PS, Canary JW, Seeman NC. Nylon/DNA: single-stranded DNA with covalently stitched nylon lining. J Am Chem Soc. 2003;125:10178–10179. doi: 10.1021/ja035186r. [DOI] [PubMed] [Google Scholar]
- 8.Zheng J, et al. 2D nanoparticle arrays show the organizational power of robust DNA motifs. Nano Lett. 2006;6:1502–1504. doi: 10.1021/nl060994c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nykypanchuk D, Maye MM, van der Lelie D, Gang O. DNA-guided crystallization of colloidal nanoparticles. Nature. 2008;451:549–552. doi: 10.1038/nature06560. [DOI] [PubMed] [Google Scholar]
- 10.Park SY, et al. DNA-programmable nanoparticle crystallization. Nature. 2008;451:553–558. doi: 10.1038/nature06508. [DOI] [PubMed] [Google Scholar]
- 11.Kim AJ, Biancaniello PL, Crocker JC. Engineering DNA-mediated colloidal crystallization. Langmuir. 2006;22:1991–2001. doi: 10.1021/la0528955. [DOI] [PubMed] [Google Scholar]
- 12.Valignat MP, Theodoly O, Crocker JC, Russel WB, Chaikin PM. Reversible self-assembly and directed assembly of DNA-linked micrometer-sized colloids. Proc Nat Acad Sci. 2005;102:4225–4229. doi: 10.1073/pnas.0500507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leunissen ME, et al. Towards self-replicating materials of DNA-functionalized colloids. Soft Matter. 2009;5:2422–2430. [Google Scholar]
- 14.Watson JD, Crick FHC. The genetical implications of the structure of deoxyribonucleic acid. Nature. 1953;171:964–67. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- 15.Lehman IR, Bessman MJ, Simms ES, Kornberg A. Enzymatic synthesis of deoxyribonucleic acids 1. J Biol Chem. 1958;233:163–170. [PubMed] [Google Scholar]
- 16.Watson JD, Crick FHC. The molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 17.Kuzuya A, Wang R, Sha R, Seeman NC. Six-helix and eight-helix DNA nanotubes assembled from half-tubes. Nano Lett. 2007;7:1757–1763. doi: 10.1021/nl070828k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang R, Kuzuya A, Liu W, Seeman NC. Blunt-ended DNA stacking interactions in a 3-helix motif. Chem Comm. 2010;46:4905–4907. doi: 10.1039/c0cc01167c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong H, Seeman NC. RNA used to control a DNA rotary machine. Nano Lett. 2006;6:2899–2903. doi: 10.1021/nl062183e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leunissen ME, et al. Switchable self-protected attractions in DNA-functionalized colloids. Nature Mater. 2009;8:590–595. doi: 10.1038/nmat2471. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z, Glotzer SC. Self-assembly of patchy particles. Nano Lett. 2004;4:1407–1413. doi: 10.1021/nl0493500. [DOI] [PubMed] [Google Scholar]
- 22.Sacanna S, Irvine WTM, Chaikin PM, Pine DJ. Lock and key colloids. Nature. 2010;464:575–578. doi: 10.1038/nature08906. [DOI] [PubMed] [Google Scholar]
- 23.Seeman NC. De novo design of sequences for nucleic acid structure engineering. J Biomol Struct & Dyns. 1990;8:573–581. doi: 10.1080/07391102.1990.10507829. [DOI] [PubMed] [Google Scholar]
- 24.Birac JJ, Sherman WB, Kopatsch J, Constantinou PE, Seeman NC. GIDEON, A program for design in structural DNA nanotechnology. J Mol Graphics & Modeling. 2006;25:470–480. doi: 10.1016/j.jmgm.2006.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.