Abstract
Arabidopsis (Arabidopsis thaliana) trichome development is a model system for studying cell development, cell differentiation, and the cell cycle. Our previous studies have shown that the GLABROUS INFLORESCENCE STEMS (GIS) family genes, GIS, GIS2, and ZINC FINGER PROTEIN8 (ZFP8), control shoot maturation and epidermal cell fate by integrating gibberellins (GAs) and cytokinin signaling in Arabidopsis. Here, we show that a new C2H2 zinc finger protein, ZFP5, plays an important role in controlling trichome cell development through GA signaling. Overexpression of ZFP5 results in the formation of ectopic trichomes on carpels and other inflorescence organs. zfp5 loss-of-function mutants exhibit a reduced number of trichomes on sepals, cauline leaves, paraclades, and main inflorescence stems in comparison with wild-type plants. More importantly, it is found that ZFP5 mediates the regulation of trichome initiation by GAs. These results are consistent with ZFP5 expression patterns and the regional influence of GA on trichome initiation. The molecular analyses suggest that ZFP5 functions upstream of GIS, GIS2, ZFP8, and the key trichome initiation regulators GLABROUS1 (GL1) and GL3. Using a steroid-inducible activation of ZFP5 and chromatin immunoprecipitation experiments, we further demonstrate that ZFP8 is the direct target of ZFP5 in controlling epidermal cell differentiation.
Cell differentiation and morphogenesis at appropriate times and places are critical for the normal development of multicellular organisms (Ishida et al., 2008). In plants, the control of cell fate is a central issue during embryo development, meristem formation, and the transition from vegetative to reproductive phases (Szymanski et al., 2000; Schellmann et al., 2007). The regulation of cell fate requires a balance of cell proliferation and differentiation, intercellular communication, and morphogenesis control. All of these developmental processes are involved in the formation of unicellular trichomes in the shoot epidermis and root hairs in the root epidermis of Arabidopsis (Arabidopsis thaliana; Szymanski et al., 2000; Pesch and Hülskamp 2009). The development of trichomes and root hairs provide excellent models for the study of the molecular basis of cell specification in plants (Szymanski et al., 2000; Schiefelbein, 2003; Pesch and Hülskamp, 2004; Schellmann et al., 2007; Wang et al., 2007). Molecular and genetic analyses have been carried out in Arabidopsis to understand the regulation of cellular differentiation programs and have shown a network of transcriptional regulators that control the development of trichome and root hair in Arabidopsis (Hülskamp et al., 1994; Szymanski et al., 2000; Ishida et al., 2008; Pesch and Hülskamp, 2009). Mutant analyses, molecular protein-protein interactions, transcriptional cross-regulatory interactions, and the movement behavior of patterning genes have shown a very complicated system for trichome development (Szymanski et al., 2000; Ishida et al., 2008; Pesch and Hülskamp, 2009; Schiefelbein et al., 2009). The integration of the transcriptional regulatory network and the movement behavior enables the simulation of de novo patterning and analysis of the robustness of the system (Benítez et al., 2007, 2008; Pesch and Hülskamp, 2009). Pesch and Hülskamp (2009) have proposed the activator-inhibitor model and the activator-depletion model to explain this complicated network regulation. The key components for the activator-depletion model are the TTG1/GLABROUS1 (GL1)/MYB23/GL3/EGL3 complex. The GL3-dependent depletion of TTG1 in trichome neighboring cells results in the formulation of an activator-depletion mechanism for trichome development (Pesch and Hülskamp, 2004, 2009). It is known that the TTG1 protein moves freely and binds to GL3 in young tissues. Consequently, the cell that has more GL3 accumulates more TTG1 and becomes more competent to develop into a trichome, whereas the neighboring cells lack TTG1 and are less competent for trichome development (Pesch and Hülskamp, 2009). To date, many GL3/GL1 direct target genes have been identified with specific expression in the very early stages of trichome initiation (Morohashi and Grotewold, 2009). The activator-inhibitor model is well supported by most researchers of trichome development in Arabidopsis.
The phytohormones GA and jasmonic acid are reported to increase trichome number and density, whereas salicylic acid decreases trichome number (Traw and Bergelson, 2003; Ishida et al., 2008). However, little information about phytohormone signaling pathways in the regulation of trichome and root hair formation is available. The first report on the involvement of GA in trichome development comes from Chien and Sussex (1996), who showed that the application of GA to the glabrous GA deficiency mutant ga1-3 induces earlier trichome formation on the adaxial epidermis compared with the abaxial epidermis and that GA stimulates trichome formation. This result was confirmed by Telfer et al. (1997), who demonstrated the promotion of trichome production in Arabidopsis by GA and that GA regulated the abaxial trichome formation and phase change. SPINDLY (SPY) encodes a repressor of GA signaling and was first cloned by Jacobsen et al. (1996), who demonstrated that mutations of SPY caused a phenotype that is consistent with constitutive activation of GA signal transduction. By using the GA-deficient mutant ga1-3, the GA-response mutant spy-5, and uniconazol (a GA biosynthesis inhibitor), Perazza et al. (1998) have shown that GA-level response correlates positively with both trichome number and trichome branch number in Arabidopsis and that GA promotes trichome formation by up-regulating GL1 in Arabidopsis. This finding was confirmed in our previous reports (Gan et al., 2006). Cytokinins also stimulate trichome formation on the inflorescence stem. GA and cytokinin signals are integrated by the C2H2 transcription factors GLABROUS INFLORESCENCE STEMS (GIS), GIS2, and ZINC FINGER PROTEIN8 (ZFP8), and they, in turn, collectively regulate GL1 expression (Gan et al., 2006, 2007a, 2007b; Ishida et al., 2008). As we have shown previously, ZFP5 is the C2H2 transcriptional factor most closely related to the GIS clade (Gan et al., 2006); thus, the aim of this research is to determine whether ZFP5 plays any role in the control of trichome development. Here, we report that a new C2H2 transcription factor, ZFP5, plays a key role in regulating inflorescence trichome development by directly targeting ZFP8 expression through a GA signaling pathway.
RESULTS
Overexpression of ZFP5 Stimulates Trichome Initiation and Causes the Heterochronic Expression of Juvenile Traits
In order to investigate whether the new C2H2 transcription factor, ZFP5, plays a role in trichome initiation, we created 35S:ZFP5 transgenic lines and found that they displayed an abnormally high density of trichomes on the second lateral branch (Fig. 1A) and inflorescence organs (Fig. 1B), which is similar to the phenotypes of 35S:GIS and 35S:ZFP8 (Gan et al., 2006, 2007b). We selected two representative transgenic lines, lines 1 and 2, which exhibited high levels of ZFP5 overexpression (Fig. 2B). Both lines had significantly more trichomes on cauline leaves, branches, and main inflorescence stems than wild-type plants (Figs. 1, A and B, and 3D). In addition, ZFP5 overexpression caused the formation of ectopic trichomes on carpels, petals, and even stamens (Fig. 1B). Furthermore, 35S:ZFP5 plants also displayed a number of phenotypic changes that we explained as heterochronic shifts in development. For example, when compared with wild-type plants, 35S:ZFP5 plants flowered significantly later than wild-type plants and exhibited more rosette leaves (Table I). Overexpression of ZFP5 also caused the occasional appearance of aerial rosettes on inflorescence stems in place of cauline leaves (Fig. 1C). In conclusion, ZFP5 overexpressors display phenotypes of a delay in shoot maturation and a strong induction of trichome production on the inflorescence.
Figure 1.
Phenotypes of loss-of-function zfp5 mutants and 35S:ZFP5 overexpressors. A, Main inflorescence stems (second lateral branch) of the wild type (WT; left), 35S:ZFP5 transgenic line 5 (second from left), zfp5 (third from left), and ZFP5:Ri (right; ZFP5 has been silenced by RNAi). B, Exposed floral organs from the wild type (left) and 35S:ZFP5 transgenic line 5 (right). The arrow points to ectopic trichomes. C, Rosette formation on the primary inflorescence stem of a strong ZFP5 overexpressor. D, Trichome initiation on the second cauline leaves of zfp5 (left) and the wild type (right). E, Trichome initiation on sepals of wild-type (left) and zfp5 (right) flowers.
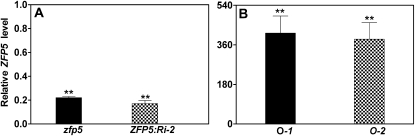
Figure 2.
Relative expression levels of ZFP5 in zfp5 and ZFP5:Ri-2 (A) and in 35S:ZFP5 lines O-1 and O-2 (B). The wild type or the corresponding control values were set at 1. lsd values were calculated at the probability of 1% (** P < 0.01).
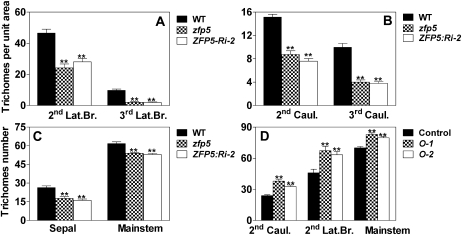
Figure 3.
Trichome initiation on inflorescence organs of zfp5, ZFP5:Ri-2, and 35S:ZFP5 lines. A to C, Trichome density on the second and third branches (A), the second and third cauline leaves (B), and sepals and main stem internodes (C) in zfp5, ZFP5:Ri-2, and wild-type (WT) plants. D, Trichome density on inflorescence organs and rosette leaves in two 35S:ZFP5 lines, O-1 and O-2. Caul., Cauline leaves; Lat.Br., lateral branch. Trichome number was calculated from at least 20 plants. Error bars represent se. lsd values were calculated at the probability of 1% (** P < 0.01).
Table I. zfp5 mutant, ZFP5:RNAi lines, and ZFP5 overexpressor affect plant development and inflorescence trichome initiation.
Flower trichome counts represent the total of at least 20 flowers; values are averages, and error bars correspond to se. The trichome number on the seventh adaxial rosette leaf is counted only at the 0.72-cm2 area in the middle of the rosette leaves. The experiment was repeated with similar results. lsd values were calculated at the probability of either 5% (* P < 0.05) or 1% (** P < 0.01). ns, Not significant; O-1, 35S:ZFP5-overexpressing line 1; O-2, 35S:ZFP5-overexpressing line 2.
| Parameter | Wild Type | zfp5 | ZFP5:Ri-2 | Control | O-1 | O-2 |
| Height at maturity (cm) | 16.66 ± 2.40 | 15.93 ± 2.25ns | 16.72 ± 1.83ns | |||
| Trichome no. on seventh adaxial rosette leaf | 17.32 ± 2.15 | 16.04 ± 1.62* | 14.87 ± 1.74** | |||
| Total trichome no. on seventh abaxial rosette leaf | 8.04 ± 2.64 | 5.04 ± 1.84** | 4.17 ± 1.56** | |||
| First leaf with abaxial trichomes | 5.96 ± 0.68 | 5.96 ± 0.61ns | 6.27 ± 0.58ns | 6.25 ± 0.55 | 8.10 ± 0.64** | 7.20 ± 0.52** |
| Total rosette leaf no. at flowering | 9.92 ± 0.89 | 10.16 ± 1.40ns | 10.30 ± 0.99ns | 12.85 ± 0.67 | 17.90 ± 1.29** | 14.65 ± 0.88** |
| Flowering time after sowing (d) | 26.64 ± 0.70 | 26.76 ± 0.66ns | 26.80 ± 0.66ns | 28.60 ± 0.60 | 34.85 ± 1.73** | 31.40 ± 1.6** |
Loss of Function of ZFP5 Affects Inflorescence Trichome Initiation
In order to understand the involvement of ZFP5 during trichome initiation, we selected a zfp5 mutant (catalog no. N583960) from the Nottingham Arabidopsis Stock Centre that carries a T-DNA insertion in the ZFP5 promoter region. The presence of T-DNA at the expected location in the zfp5 mutant was verified by genomic PCR. We also constructed the vector in which ZFP5 was silenced by RNA interference (RNAi; line ZFP5:Ri-2). We have generated at least six lines for ZFP5:RNAi, and all of them showed similar phenotypes. Quantitative PCR analysis showed that ZFP5 expression was significantly suppressed in the zfp5 mutant and ZFP5:Ri-2 (Fig. 2A). To further characterize the pattern of trichome initiation in the zfp5 mutant, we analyzed in detail the trichome distribution on stems, lateral branches, cauline leaves, and flowers in the zfp5 mutant, ZFP5:Ri-2, and the control wild-type plants. We found that trichome initiation in the zfp5 mutant was not affected on the first lateral branch, the first cauline leaf, and the first internode but was significantly decreased on successive stem internodes, branches, cauline leaves, and sepals (Figs. 1, D and E, and 3, A–C).
ZFP5 Controls Trichome Shoot Maturation through GA Signaling
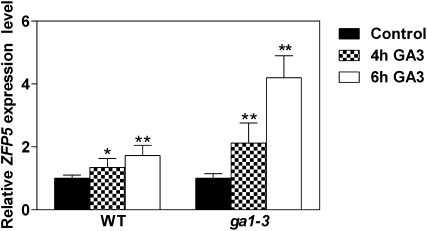
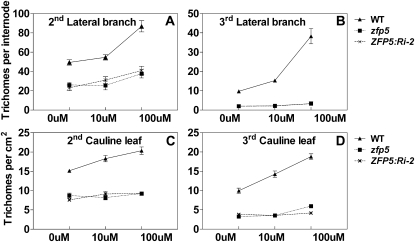
In order to study whether ZFP5 controls trichome initiation through GA signaling like GIS, we examined whether the expression level of ZFP5 in inflorescence organs of the wild type was induced by external GA application. Wild-type and ga1-3 mutant plants were treated with 100 μm GA, and plants were harvested at 4 and 6 h after GA application. As shown in Figure 4, ZFP5 expression was significantly higher in GA-treated plants than in mock-treated plants in both wild-type and ga1-3 backgrounds. This result suggests that ZFP5 expression is induced by GA. GA is known to promote trichome initiation on leaves and inflorescence organs (Chien and Sussex, 1996; Gan et al., 2006). In order to study the response of the zfp5 mutant to external GA application, wild-type, zfp5, and ZFP5:RNAi-2 transgenic lines were treated with 10 and 100 μm GA shortly after flowering, as described before (Gan et al., 2006). The results showed that trichome initiation was less sensitive (the second lateral branch) than the wild type or completely insensitive (the third lateral branch and cauline leaves) to GA application in zfp5 and ZFP5:RNAi-2. These results indicate that ZFP5 is required for the GA signaling pathway in stimulating trichome initiation on inflorescence organs (Fig. 5, A and B).
Figure 4.
Expression of ZFP5 in response to GA application. Expression of ZFP5 was measured in inflorescence organs of wild-type (WT) and gal-3 plants after GA treatment (100 μm) for 4 and 6 h. Transcript levels were measured by real-time PCR, and the values were normalized against the levels of UBQ10 as a control. lsd values were calculated at the probability of either 5% (* P < 0.05) or 1% (** P < 0.01).
Figure 5.
The effect of GA application on trichome initiation in wild-type (WT), zfp5 mutant, and ZFP5:RNAi-2 transgenic lines. Trichome initiation is shown on the inflorescence organs of wild-type, zfp5, and ZFP5:RNAi (termed ZFP5:Ri-2) plants that were treated with different concentrations of GA. Error bars indicate se. The second and third lateral branches and the second and third cauline leaves were examined from 20 plants.
ZFP5 Acts Upstream of the Trichome Initiation Complex
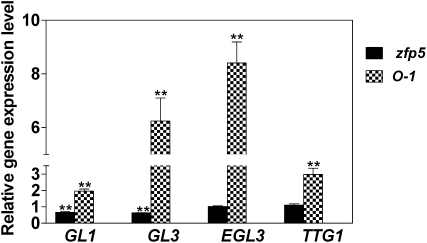
Trichome initiation is controlled by the TTG1/GL1/MYB23/GL3/EGL3 complex, GIS, and its subfamily genes GIS2 and ZFP8, which act downstream of SPY and upstream of GL1. To investigate the genetic position of ZFP5 in the trichome initiation pathway, we examined the relative expression levels of GL1, GL3, EGL3, and TTG1 in developing inflorescence shoots of the zfp5 mutant and 35S:ZFP5. We found that expression of GL1, GL3, and EGL3 was significantly down-regulated in the zfp5 mutant but up-regulated in 35S:ZFP5 (Fig. 6). We further overexpressed ZFP5 in the backgrounds of gl1, gl3, and ttg1 and revealed that the glabrous phenotype of these mutants was not rescued by overexpression of ZFP5 (Supplemental Fig. S1, A and B). However, overexpression of a GL3/EGL3 homologous R gene was able to rescue the zfp5 mutant phenotype (Supplemental Fig. S1C). These results support that, like GIS, ZFP5 acts upstream of GL1 and GL3/EGL3.
Figure 6.
Relative expression of GL1, GL3, EGL3, and TTG1 in developing inflorescence shoots of zfp5 and 35S:ZFP5. Values represent the ratios of gene expression in a particular genotype to the corresponding wild type or transgenic control. O-1, 35S:ZFP5-overexpressing line 1. lsd values were calculated at the probability of 1% (** P < 0.01).
ZFP5 Acts Upstream of GIS, GIS2, and ZFP8
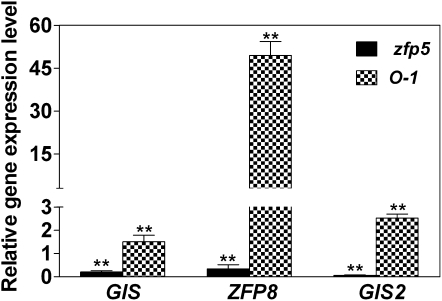
Since the zfp5 mutant showed a similar phenotype to GIS family mutants, the genetic position between ZFP5 and GIS family genes was investigated. We examined the expression of GIS, ZFP8, and GIS2 in zfp5 and 35S:ZFP5. We found that relative expression of GIS, ZFP8, and GIS2 in the developing inflorescence shoots of zfp5 mutants was significantly lower than in the wild-type plants but significantly higher in 35S:ZFP5 than in wild-type plants, particularly for the expression of ZFP8 in 35S:ZFP5 (Fig. 7). Further investigation of the expression level of ZFP5 in developing inflorescence shoots of gis, gis2, and zfp8 mutants revealed that the expression of ZFP5 in these mutants was not significantly different from that in wild-type plants (Supplemental Fig. S2). This result suggests that ZFP5 functions upstream of GIS, GIS2, and ZFP8. To further confirm these results, we overexpressed ZFP8 in the background of zfp5 and revealed that the phenotype of the zfp5 mutant was rescued by overexpression of ZFP8 (Fig. 8). This results also suggests that ZFP5 acts upstream of ZFP8.
Figure 7.
Relative expression of GIS, ZFP8, and GIS2 in developing inflorescence shoots of zfp5 and 35S:ZFP5 (O-1). The wild-type or the corresponding control values were set at 1. lsd values were calculated at the probability of 1% (** P < 0.01).
Figure 8.
Overexpressed ZFP8 restores ZFP5 mutant trichome phenotype. A, Trichome production on cauline leaves of the wild type (WT; left), 35S:ZFP8:zfp5 (center), and zfp5 (right). B, Trichome initiation on the second branch of the wild type (left), 35S:ZFP8:zfp5 (center), and zfp5 (right). [See online article for color version of this figure.]
ZFP8 Is the Direct Target of ZFP5
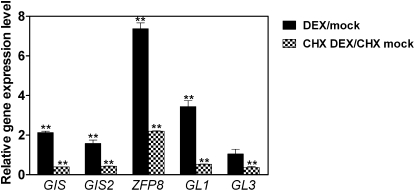
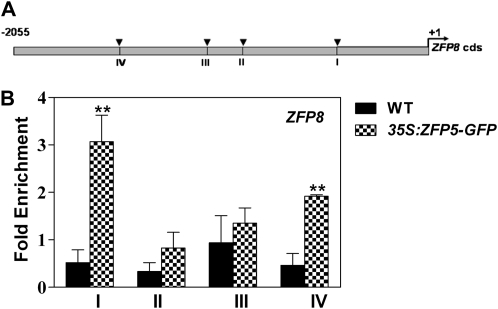
To further elucidate whether GIS, GIS2, ZFP8, GL1, and GL3 are the downstream targets of ZFP5, we created a steroid-inducible version of ZFP5 in transgenic plants containing the ZFP5 protein fused to the hormone-binding domain of a rat GR under the control of a 35S promoter. Posttranslational activation of ZFP5 can be achieved by dexamethasone (DEX) treatment of the resulting transgenic plants (Wagner et al., 1999; Yu et al., 2004). In these plants, ZFP5 (35S:ZFP5-GR) activity is inducible by DEX. To validate our results, we analyzed gene expression in response to 35S:ZFP5-GR activity in the zfp5 mutant background. The effects of 35S:ZFP5-GR induction were determined at different time points following DEX treatment. The results show that the expression of the key trichome initiation-positive regulators GIS, GIS2, ZFP8, GL1, and GL3 is rapidly induced and that effect was detectable as early as 2 h after DEX application (Fig. 9). In the case of ZFP8, the induction was the highest, with a more than 7-fold increase. Because DEX induction of 35S:ZFP5-GR activity does not require protein synthesis, it is possible to determine whether its effects are direct or indirect by examining the influence of protein synthesis inhibitors on induced gene expression changes (Gan et al., 2007a). To determine whether 35S:ZFP5-GR directly or indirectly promotes trichome regulator expression, we repeated DEX applications to 35S:ZFP5-GR zfp5 in the presence of the protein synthesis inhibitor cycloheximide. We found that expression of ZFP8 is still induced by DEX in the presence of cycloheximide, which suggests that ZFP8 could be the direct target of ZFP5. Although 35S:ZFP5-GR activity rapidly triggers the induction of trichome initiation activators GIS, GIS2, ZFP8, GL1, and GL3, cycloheximide treatment strongly inhibits the induction of GIS, GIS2, GL1, and GL3, which suggests that they are not the direct target of ZFP5 (Fig. 9). To further confirm whether ZFP8 is directly regulated by ZFP5, we performed a chromatin immunoprecipitation (ChIP) assay using 35S:ZFP5-GFP transgenic plants to test whether ZFP5 could bind directly to the promoter of ZFP8. We scanned a length of 2,055-bp ZFP8 promoter and located four fragments spanning the conserved sequence A[AG/CT]CNAC (Fig. 10A), which were considered the C2H2 zinc finger protein-binding site of their target genes (Sakai et al., 1995; Kubo et al., 1998). Our results show that fragments I and IV are readily enriched with GFP antibody (Fig. 10B). These data further support the hypothesis that ZFP5 is directly targeted to ZFP8.
Figure 9.
Induced ZFP5 activity can transcriptionally activate the expression of GIS, GIS2, ZFP8, GL1, and GL3, and ZFP8 is the direct target of ZFP5. Quantitative RT-PCR analysis is shown for RNA from the zfp5 mutant harboring ZFP5: GR treated with 10 μm DEX for 2 h (black bars) and treated with 10 μm DEX and 20 μm CHX for 2 h (hatched bars). Values are averages, and error bars correspond to se. lsd values were calculated at the probability of 1% (** P < 0.01).
Figure 10.
ChIP analysis of ZFP8 promoter regions bound by 35S:ZFP5-GFP. A, Schematic diagram of the ZFP8 promoter. The arrowheads indicate the sites containing either a single mismatch or a perfect match from the consensus binding sequence A[AG/CT]CNAC for C2H2 zinc finger proteins. B, Quantitative real-time PCR assay of DNAs after ChIP. TUBULIN2 was used as a reference, and the primer used was described by Yu et al. (2010). Values are averages, and error bars correspond to se. lsd values were calculated at the probability of 1% (** P < 0.01). WT, Wild type.
ZFP5 Belongs to a Subfamily of the GIS C2H2 Gene Family That Is Highly Expressed in Root and at Early Stages of Inflorescence Development
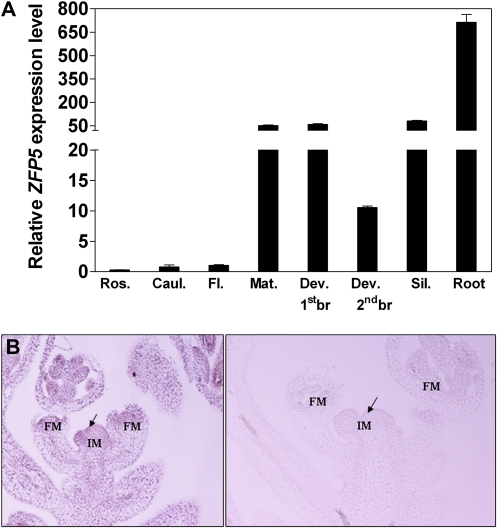
Sequence analysis showed that ZFP5 contains a C2H2 domain found in transcription factors of the TFIIIA class and is most similar to the ZFP6 and GIS family of transcription factors (Meissner and Michael, 1997; Payne et al., 2004; Gan et al., 2006, 2007b). Quantitative reverse transcription (RT)-PCR analysis of ZFP5 expression showed that ZFP5 had a different expression pattern to that of GIS and its family genes GIS2 and ZFP8, which were hardly detected in roots. ZFP5 was highly expressed in roots, developing stems, branches, and young leaves but was low in rosette leaves and mature cauline leaves (Fig. 11A). We further performed in situ hybridization using a ZFP5-specific probe on sections of developing inflorescence stems and revealed that the gene was widely expressed in the inflorescence apex (Fig. 11B).
Figure 11.
Expression pattern of the ZFP5 gene. A, Quantitative RT-PCR analysis of ZFP5 expression in different tissues of wild-type plants. All values are normalized using an internal standard (UBQ10). Ros., Rosette leaf; Caul., cauline leaf; Fl., flower; Mat. stem, fully elongated first internode of the main stem; Dev. 1stbr., the first developing branch (lateral branch); Dev. 2ndbr., the second developing branch (lateral branch); Sil., silique. B, In situ hybridization of ZFP5 probes to developing inflorescence shoot sections. ZFP5 is expressed broadly in stems and inflorescence meristems (IM) and strongly in floral meristems (FM). The section hybridized with the antisense probe is shown on the left, while the one hybridized with the sense probe is shown on the right.
DISCUSSION
Functional Specialization of ZFP5
The surface of aerial organs in plants typically produce trichomes that have various functions against biotic and abiotic stresses in a variety of plant species, which include tobacco (Nicotiana tabacum), tomato (Solanum lycopersicum), soybean (Glycine max), and pepper (Capsicum annuum; Du et al., 2009; Harada et al., 2010; Kang et al., 2010; Kim et al., 2010). Our previous findings show that GIS and its related family genes GIS2 and ZFP8 play different roles in regulating epidermal differentiation on different aerial organs in Arabidopsis (Gan et al., 2006, 2007a). GIS, ZFP8, and GIS2 have diverged in their responses to both developmental and hormonal signals, and these signals are not strictly interdependent (Gan et al., 2007b). The roles of GIS, ZFP8, and GIS2 appear to be mainly in the modulation of specific epidermal responses to cytokinins and GAs. GIS plays a key role in controlling trichome initiation on inflorescence stems, GIS2 mainly controls trichome initiation on inflorescence flowers, while ZFP8 is the key regulator for trichome initiation on cauline leaves (Gan et al., 2006, 2007b). As we reported before, although the GIS, ZFP8, and GIS2 genes are functionally equivalent proteins, they have diverged in their responses to plant hormones and in their roles during inflorescence development (Gan et al., 2006, 2007b). As the expression patterns of GIS, ZFP8, and GIS2 are different and both the roles and inducibility of the genes in response to cytokinins are clearly different, we suggest that GIS, ZFP8, and GIS2 have diverged in their responses to both developmental and hormonal signals and that these signals are not strictly interdependent (Gan et al., 2007b). Thus, there could be two different pathways: one where GIS controls trichome development mainly by the regulation of expression of GL1/GL3 (Gan et al., 2006, 2007b) through GA, and another, the GIS2/ZFP8 pathway, which integrates both GA and cytokinin signals to control trichome development in inflorescence organs (Gan et al., 2007b).
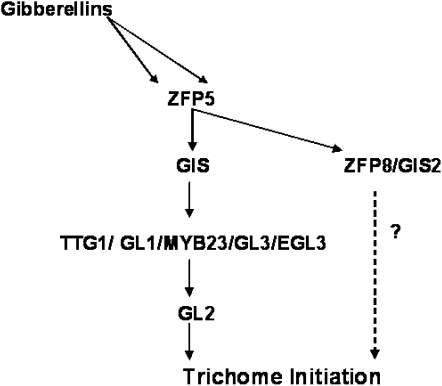
In this study, ZFP5 differs from the GIS clade, which plays a general role in modulating growth and differentiation in response to GA signals (Achard et al., 2003, 2006; Fu and Harberd, 2003; Gan et al., 2006, 2007a, 2007b). It appears that ZFP5 plays a predominant role in trichome initiation on inflorescence stems and cauline leaves (Figs. 1, D and E, and 3, B and C). Our genetic and molecular analyses have shown that ZFP5 acts upstream of the trichome key positive regulators GL1, GL3, GIS, GIS2, and ZFP8 by directly targeting ZFP8 (Fig. 10). Therefore, ZFP5 highlights the key role played by transcription factors not only upstream of the GIS clade but also downstream of hormone production (Fig. 12). The model of ZFP5 controlling epidermal cell differentiation is summarized in Figure 12. These results will provide a better understanding of how developmental and hormonal signals are integrated in plants by transcription factors. A recent report from Yu et al. (2010) demonstrates that the SQUAMOSA PROMOTER-BINDING PROTEIN-LIKE gene SPL9, which is targeted by microRNA156 (miR156), plays a key role in controlling inflorescence trichome development by directly targeting TRY and TCL1. Yu et al. (2010) further demonstrated that miR156/SPL is not related to the GIS-dependent pathway and that these two pathways function in parallel in regulating the trichome regulator complex in the main stem and inflorescences. Since ZFP5 belongs to a subfamily gene of the GIS C2H2 family and acts upstream of the GIS gene family by directly regulating ZFP8, it is unlikely to play any role in the miR156/SPL-dependent pathway. Since our gene expression data have shown clearly that ZFP5 is highly expressed in the root, further study is needed to determine whether ZFP5 plays a role in the control of root or root hair development.
Figure 12.
Model of ZFP5 action in Arabidopsis inflorescence organs in response to GA.
ZFP5 Regulates Phase Change
As we reported before, the heterochronic phenotype of GIS, GIS2, and ZFP8 overexpressors and loss-of-function mutants suggested that GIS, GIS2, and ZFP8 influence trichome initiation as part of a more general role in the regulation of phase change (Gan et al., 2006, 2007b). In order to test whether ZFP5 plays a role in this phase change, we first investigated the phenotype resulting from the overexpression of ZFP5. The results showed that the ZFP5 overexpressor delays flowering time and the transition period from vegetative stage to reproductive stage (Table I), which suggests that ZFP5 may play a general role in regulating phase change. We further examined the process of shoot maturation in the zfp5 mutant. We found that the mutation did not affect flowering time in long days or the adaxial leaf number, but it significantly affected the abaxial leaf number of the rosette leaves (Table I). These results suggested that a loss of ZFP5 function not only affects epidermal differentiation after floral induction under normal growth conditions but also plays a role in phase change. Elucidating how increases in GIS activity and its family genes influence phase change will require further investigation.
In summary, we have cloned and characterized a new C2H2 zinc finger protein, ZFP5, which plays a key role in shoot maturation and trichome initiation in the inflorescence organs. Our results clearly demonstrated that ZFP5 controls shoot maturation by regulating the expression of GIS, GIS2, ZFP8, GL1, and GL3 through integrating GA signaling by directly targeting ZFP8 (Fig. 10). This is another big step for us to better understand the transcriptional factor controlling trichome initiation by integrating GA signal. Since there are two more genes, ZFP6 and At1g68360 (Gan et al., 2006), in the ZFP5 clade, the cloning and characterization of these two genes should bring further insights into this mode of action for the transcriptional factors controlling shoot maturation and trichome cell differentiation by integrating plant hormones and environmental cues.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used as a control for most experiments in this study. The plants were grown in a controlled growth room with the following growth conditions: 20°C to 22°C, 90 to 120 μmol m−2 s−1, 16/8-h photoperiod, and 68% to 78% humidity. For in the vitro experiments, seeds were surface sterilized with 5% (v/v) NaClO solution for 7 min and washed five times in sterile distilled water, plated on Murashige and Skoog medium and vernalized at 4°C for 3 d in the dark, and then placed in a growth room.
Isolation of a zfp5 Loss-of-Function Mutant
A transgenic line (catalog no. N583960) carrying a T-DNA insertion in the ZFP5 promoter was identified in the Nottingham Arabidopsis Stock Centre line collection. Homozygous mutants were selected by ensuring that all of their progeny were resistant to kanamycin (50 mg L−1). The presence of the T-DNA insertion was confirmed by PCR using gene-specific primers (5′ primer, 5′-TGTGACGAGAAATGATCTTGG-3′; 3′ primer, 5′-ATAGATGCTTGCATCATTGCC-3′) and a T-DNA insertion primer (5′-ATTTTGCCGATTTCGGAAC-3′).
Cloning
For all cloning constructs (35S:ZFP5, ZFP5:RNAi,), sequences were first inserted into the pENTR-1A vector (Invitrogen) before being recombined into an appropriate destination vector using the Gateway LR reaction (Invitrogen). All destination vectors were obtained from the Flanders Interuniversity Institute for Biotechnology as described before (Gan et al., 2006). pH2GW7 was used for preparing the 35S:ZFP5 construct, pK7GWIWG2(II), for the ZFP5:RNAi construct. For all these cloning experiments, gene-specific fragments were first PCR amplified from cDNA (35S:ZFP5, ZFP5:RNAi) using primers containing SalI and NotI restriction sites and purified using a gel extraction kit (Qiagen) before restriction and cloning. The following primers were used: ZFP5 overexpression, 5′-CGGTCGACATGTCTATAAATCCG-3′ and 5′-AAGCGGCCGCTGCCGCATCTCCGGCA-3′; ZFP5:RNAi, 5′-CGGTCGACGAGCCAGCATCGGCTATTAT-3′ and 5′-AAGCGGCCGCAATTTGTTATGCCGCATCTCC-3′. For all of the genetic transgenic experiments, the constructs in binary vectors were transformed into Agrobacterium tumefaciens GV3101 strain, which were then used to transform Arabidopsis with the appropriate genotype background using the floral-dip method as described by Clough and Bent (1998). Transgenic seedlings were first selected on Murashige and Skoog agar plates with appropriate antibiotics and then confirmed by PCR using the corresponding primers. We created 35S:ZFP5-GR using a derivative pGreen0244 vector (pGreen0244:GR donated by Prof. Hao Yu). The entire ZFP5 cDNA was amplified by RT-PCR with the primers ZFP5-LP (5′-CACTACGCGTATGTCTATAAATCCGACAAT-3′) and ZFP5-RP (5′-TAATGGGCCCTGCCGCATCTCCGGCAAAAC-3′). The resulting fragment was digested by ApaI and MluI sites of pGreen0244 and subsequently cloned into the corresponding sites of pGreen0244 to generate 35S:ZFP5-GR.
GA Treatments and Gene Expression Analysis
GA3 (Sigma) was used in all experiments that involved exogenous GA treatments, as described previously (Gan et al., 2006, 2007b). For measuring the effect of GA applications on gene expression, a minimum of eight mutant and control plants were grown on soil until young inflorescence shoots had reached a size of 2 to 3 cm. The plants were then sprayed with either 100 μm GA3 or a mock solution, and the shoots were harvested 4 and 6 h (GA) after treatment for RNA extraction (Gan et al., 2006).
RNA Extraction and Real-Time PCR
The total RNA was extracted from Arabidopsis organs with TRIzol Reagent (Invitrogen), and cDNA was synthesized from 1.5 μg of total RNA using Moloney murine leukemia virus transcriptase (Promega) and oligo(dT)18 primers in a 25-μL reaction according to the manufacturer’s instructions. For real-time PCR, the cDNAs were diluted to 100 μL, and 2 μL was added to 12.5 μL of SYBR Green PCR mix (Takara) and 0.5 μL of each primer (200 nm final concentration) in 25-μL reactions. PCR and detection were performed using a Stratagene M×3005P thermal cycler using the following cycling conditions: 95°C for 1 min, followed by 40 cycles of 95°C for 5 s and 60°C for 20 s. Optimization experiments were performed to establish the optimal PCR programs. Melting-curve analysis was used to confirm the absence of nonspecific amplification products. UBQ10 transcripts were used as an endogenous control to normalize expression of the other genes according to Gan et al. (2006). Relative expression levels were calculated by subtracting the threshold cycle (Ct) values for UBQ10 from those of the target gene (to give ΔCt) and then calculating 2−ΔCt as described previously (Gan et al., 2005). RT-PCR experiments were performed on at least three independent samples.
In Situ Hybridization
Nonradioactive in situ hybridization was performed as described previously (Gan et al., 2006). For synthesis of the ZFP5 RNA probes, gene-specific fragments were amplified using the same primers as for generating the RNAi constructs (see above).
DEX Treatment
For the 35S:ZFP5-GR transgenic line selection, we screened for a transgenic line that contains only one transgene insertion and that exhibits a phenotype closely resembling 35S:ZFP5. The DEX treatment and sample collection were the same as described by Yu et al. (2004) and Wagner et al. (1999). Relative expression levels were calculated according to Yi et al. (2010). UBQ10 transcripts were used as an endogenous control to normalize expression of the other genes. Quantitative RT-PCR experiments were performed at least on three independent samples.
ChIP
ChIP assays were carried out as described by Yu et al. (2010). About 3 g of tissues of 4-week-old 35S:ZFP5-GFP plants and wild-type plants were harvested and fixed in 15.4 mL of fixation buffer (0.4 m Suc, 10 mm Tris, pH 8.0, 1 mm EDTA, pH 7.5, 1 mm phenylmethylsulfonyl fluoride, and 1% formaldehyde). After fixation, the material was washed four times with double distilled water and ground under liquid nitrogen. The resultant powder was added to 10 mL of extraction buffer (0.4 m Suc, 10 mm Tris-HCl, pH 8.0, 10 mm MgCl2, 5 mm mercaptoethanol, 0.1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor [Roche]), vortexed to mix, kept at 4°C until the solution was homogenous, and then rotated for 20 min at 4°C. After that, the pellet was resuspended with lysis buffer (50 mm HEPES, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 0.1% deoxycholate sodium, and 0.1% SDS) and sonicated with an ultrasonic cell disruptor (JY96-II [output 3 and 6 × 10 s]; Xinzhi). The solution was divided into three parts: one was saved as total DNA without precipitation, and other two parts were incubated with either an anti-GFP antibody (Abmart) or an anti-hemagglutinin antibody (Abmart) , the latter of which was precipitated in parallel as a negative control. The resulting DNA samples were purified with a PCR purification kit (Qiagen). The relative concentrations of the DNA fragments were analyzed by quantitative real-time PCR using the β-TUBULIN2 gene promoter as the reference, and DNA enrichment was examined by quantitative real-time PCR in triplicate as described previously (Jun et al., 2010; Yu et al., 2010). All primers used in the ChIP assays are listed in Table II.
Table II. Primer pairs used for ChIP assays.
| Gene | Sequence (5′–3′) | Purpose |
| ZFP8 chip LP1 | TCCTATGCTTCCATACGCTAA | ChIP, region I |
| ZFP8 chip RP1 | TGACTCCATGGTGCATGTTT | ChIP, region I |
| ZFP8 chip LP2 | AACCAAATAGGTCCGTGATGTT | ChIP, region II |
| ZFP8 chip RP2 | AGCAGATGAAATGGGGTCAC | ChIP, region II |
| ZFP8 chip LP3 | TCATGAACATGGACCACCTC | ChIP, region III |
| ZFP8 chip RP3 | GGATGTTTTGTCTCTTTGATGC | ChIP, region III |
| ZFP8 chip LP4 | CCGCTCAAAACAACTAACTGAA | ChIP, region IV |
| ZFP8 chip RP4 | TTTGACGACTCCCAAATTCC | ChIP, region IV |
| TUB2 chip LP | TAAATTCTGAACCCATTGTTTCTCA | Control |
| TUB2 chip RP | AGTCCGATGATTGGCTTTATTATTC | Control |
Statistical Analyses
All data in Table I were tested by means of ANOVA for significance by using the Statistix program version 3.5 (Analytical Software). lsd values were calculated at either 5% (P < 0.05) or 1% (P < 0.01) probability as we described before (Gan et al., 2010).
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: ZFP5 (At1g10480), GIS (At3g58070), GIS2 (At5g06650), ZFP8 (At2g41940), GL1 (At3g27920), GL3 (At5g41315), EGL3 (At1g63650), TTG1 (At5g24520), TUB2 (At5g62690), and UBQ10 (At4g05320).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Genetic interaction between ZFP5 and key trichome initiation complex genes.
Supplemental Figure S2. ZFP5 expression in developing inflorescence shoots of gis, zfp8, and gis2.
Acknowledgments
We thank Dr. Clare Steele-King (University of York) for critical reading of the manuscript and Prof. Alan Lloyd (University of Texas at Austin) for providing the gl3-1 mutant. Technical assistance and help with the ChIP experiment from Dr. Nan Yu and Dr. Wenjuan Cai of Prof. Xiaoya Chen’s laboratory (Shanghai Institute for Biological Science) are gratefully acknowledged.
References
- Achard P, Cheng H, De Grauwe L, Decat J, Schoutteten H, Moritz T, Van Der Straeten D, Peng J, Harberd NP. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Achard P, Vriezen WH, Van Der Straeten D, Harberd NP. (2003) Ethylene regulates Arabidopsis development via the modulation of DELLA protein growth repressor function. Plant Cell 15: 2816–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benítez M, Espinosa-Soto C, Padilla-Longoria P, Alvarez-Buylla ER. (2008) Interlinked nonlinear subnetworks underlie the formation of robust cellular patterns in Arabidopsis epidermis: a dynamic spatial model. BMC Syst Biol 2: 98–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benítez M, Espinosa-Soto C, Padilla-Longoria P, Diaz J, Alvarez-Buylla ER. (2007) Equivalent genetic regulatory networks in different contexts recover contrasting spatial cell patterns that resemble those in Arabidopsis root and leaf epidermis: a dynamic model. Int J Dev Biol 51: 139–155 [DOI] [PubMed] [Google Scholar]
- Chien JC, Sussex IM. (1996) Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol 111: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Du WJ, Yu DY, Fu SX. (2009) Analysis of QTLs for the trichome density on the upper and downer surface of leaf blade in soybean Glycine max (L.) Merr. Agric Sci China 8: 529–537 [Google Scholar]
- Fu X, Harberd NP. (2003) Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421: 740–743 [DOI] [PubMed] [Google Scholar]
- Gan Y, Filleur S, Rahman A, Gotensparre S, Forde BG. (2005) Nutritional regulation of ANR1 and other root-expressed MADS-box genes in Arabidopsis thaliana. Planta 222: 730–742 [DOI] [PubMed] [Google Scholar]
- Gan Y, Kumimoto R, Liu C, Ratcliffe OJ, Yu H, Broun P. (2006) GLABROUS INFLORESCENCE STEMS modulates the regulation by gibberellins of epidermal differentiation and shoot maturation in Arabidopsis. Plant Cell 18: 1383–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y, Yu H, Peng J, Broun P. (2007a) Genetic and molecular regulation by DELLA proteins of trichome development in Arabidopsis. Plant Physiol 145: 1031–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan Y, Zhou Z, An L, Bao S, Liu Q, Srinivasa M, Goddard P. (2010) The effects of fluctuations in the nutrient supply on the expression of ANR1 and 11 other MADS box genes in shoots and roots of Arabidopsis thaliana. Botany 88: 1023–1031 [Google Scholar]
- Gan YB, Liu C, Yu H, Broun P. (2007b) Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development 134: 2073–2081 [DOI] [PubMed] [Google Scholar]
- Harada E, Kim JA, Meyer AJ, Hell R, Clemens S, Choi YE. (2010) Expression profiling of tobacco leaf trichomes identifies genes for biotic and abiotic stresses. Plant Cell Physiol 51: 1627–1637 [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Misŕa S, Jürgens G. (1994) Genetic dissection of trichome cell development in Arabidopsis. Cell 76: 555–566 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T. (2008) A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365–386 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE. (1996) SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci USA 93: 9292–9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JH, Ha CM, Fletcher JC. (2010) BLADE-ON-PETIOLE1 coordinates organ determinacy and axial polarity in Arabidopsis by directly activating ASYMMETRIC LEAVES2. Plant Cell 22: 62–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JH, Liu GH, Shi F, Jones AD, Beaudry RM, Howe GA. (2010) The tomato odorless-2 mutant is defective in trichome-based production of diverse specialized metabolites and broad-spectrum resistance to insect herbivores. Plant Physiol 154: 262–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Han JH, Kwon JK, Park M, Kim BD, Choi D. (2010) Fine mapping of pepper trichome locus 1 controlling trichome formation in Capsicum annuum L. CM334. Theor Appl Genet 120: 1099–1106 [DOI] [PubMed] [Google Scholar]
- Kubo K, Sakamoto A, Kobayashi A, Rybka Z, Kanno Y, Nakagawa H, Takatsuji H. (1998) Cys2/His2 zinc-finger protein family of petunia: evolution and general mechanism of target-sequence recognition. Nucleic Acids Res 26: 608–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner R, Michael AJ. (1997) Isolation and characterisation of a diverse family of Arabidopsis two and three-fingered C2H2 zinc finger protein genes and cDNAs. Plant Mol Biol 33: 615–624 [DOI] [PubMed] [Google Scholar]
- Morohashi K, Grotewold E. (2009) A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet 5: e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne T, Johnson SD, Koltunow AM. (2004) KNUCKLES (KNU) encodes a C2H2 zinc-finger protein that regulates development of basal pattern elements of the Arabidopsis gynoecium. Development 131: 3737–3749 [DOI] [PubMed] [Google Scholar]
- Perazza D, Vachon G, Herzog M. (1998) Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis. Plant Physiol 117: 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M, Hülskamp M. (2004) Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Curr Opin Genet Dev 14: 422–427 [DOI] [PubMed] [Google Scholar]
- Pesch M, Hülskamp M. (2009) One, two, three...models for trichome patterning in Arabidopsis? Curr Opin Plant Biol 12: 587–592 [DOI] [PubMed] [Google Scholar]
- Sakai H, Medrano LJ, Meyerowitz EM. (1995) Role of SUPERMAN in maintaining Arabidopsis floral whorl boundaries. Nature 378: 199–203 [DOI] [PubMed] [Google Scholar]
- Schellmann S, Hülskamp M, Uhrig J. (2007) Epidermal pattern formation in the root and shoot of Arabidopsis. Biochem Soc Trans 35: 146–148 [DOI] [PubMed] [Google Scholar]
- Schiefelbein J. (2003) Cell-fate specification in the epidermis: a common patterning mechanism in the root and shoot. Curr Opin Plant Biol 6: 74–78 [DOI] [PubMed] [Google Scholar]
- Schiefelbein J, Kwak SH, Wieckowski Y, Barron C, Bruex A. (2009) The gene regulatory network for root epidermal cell-type pattern formation in Arabidopsis. J Exp Bot 60: 1515–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski DB, Lloyd AM, Marks MD. (2000) Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci 5: 214–219 [DOI] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS. (1997) Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645–654 [DOI] [PubMed] [Google Scholar]
- Traw MB, Bergelson J. (2003) Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol 133: 1367–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner D, Sablowski RW, Meyerowitz EM. (1999) Transcriptional activation of APETALA1 by LEAFY. Science 285: 582–584 [DOI] [PubMed] [Google Scholar]
- Wang SC, Kwak SH, Zeng QN, Ellis BE, Chen XY, Schiefelbein J, Chen JG. (2007) TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 134: 3873–3882 [DOI] [PubMed] [Google Scholar]
- Yi K, Menand B, Bell E, Dolan L. (2010) A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat Genet 42: 265–267 [DOI] [PubMed] [Google Scholar]
- Yu H, Ito T, Zhao Y, Peng J, Kumar P, Meyerowitz EM. (2004) Floral homeotic genes are targets of gibberellin signaling in flower development. Proc Natl Acad Sci USA 101: 7827–7832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Cai WJ, Wang SC, Shan CM, Wang LJ, Chen XY. (2010) Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell 22: 2322–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]