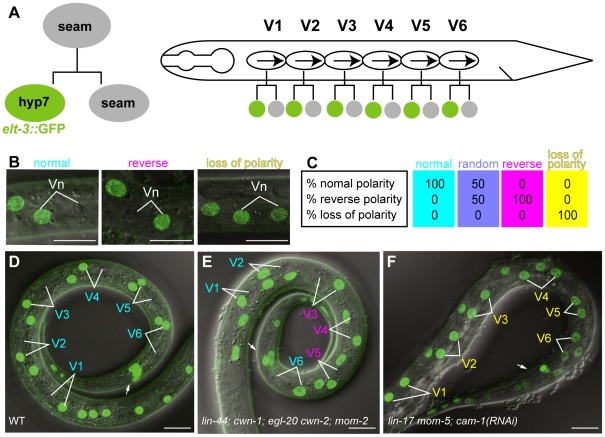
Figure 1. Analyses of seam cell polarity.
(A) Schematic drawing of seam cell divisions. Left: Anterior daughter of a seam cell fuses with hyp7 and becomes elt-3::GFP-positive. Right: seam cells (V1–V6) on the lateral sides of animals are polarized in the same orientation (represented by arrows) and divide asymmetrically. (B) Examples of the polarity of seam cell (Vn) divisions (normal, reverse, or loss of polarity) as judged by the elt-3::GFP expression in the daughter cells. Merged differential interference contrast (DIC) and fluorescent images are shown. (C) An illustration of sample data for the polarity of seam cell divisions for various genotypes. Each colored box represents the polarity of an individual seam cell. Top, middle, and bottom numbers in the boxes are the percentages of individual seam cells with normal, reverse, and loss of polarity, respectively. An RGB color component was assigned for each polarity phenotype (normal = red; reverse = green; loss of polarity = blue), and the color of each box represents the combination of the calculated intensity of each RGB component per seam cell. Intensity was calculated as follows: 255−(% observed phenotype×2.55), where 255 is the maximum intensity of each RGB component, and 2.55 is a factor for standardizing the phenotype percentage to the RGB scale. The colored boxes shown here represent the resulting four standard colors. Cells with 100% normal division-polarity are cyan (red 0, green 255, blue 255); cells with random division-polarity are lavender (red 128, green 128, blue 255); cells with 100% reverse division-polarity are magenta (red 255, green 0, blue 255); and cells with 100% loss of division-polarity are yellow (red 255, green 255, blue 0). In the similar illustrations in the following Figures, intermediate color compositions indicate relative tendencies towards a certain phenotype in relation to these four standards. (D–F) The expression of elt-3::GFP at the late L1 stage in wild-type C. elegans (D); lin-44(n1792); cwn-1(ok546); egl-20(n585) cwn-2(ok895); mom-2(ne874ts); vpIs1 (E); and lin-17(n3091) mom-5(ne12); cam-1(RNAi); vpIs1 (F). The colors of the cell names indicate the polarity of their divisions (normal in cyan, reverse in magenta, and loss of polarity in yellow), as determined by the elt-3::GFP expression in their daughter cells. Arrows indicate the anus, which is on the ventral side. Scale bars: 10 µm.