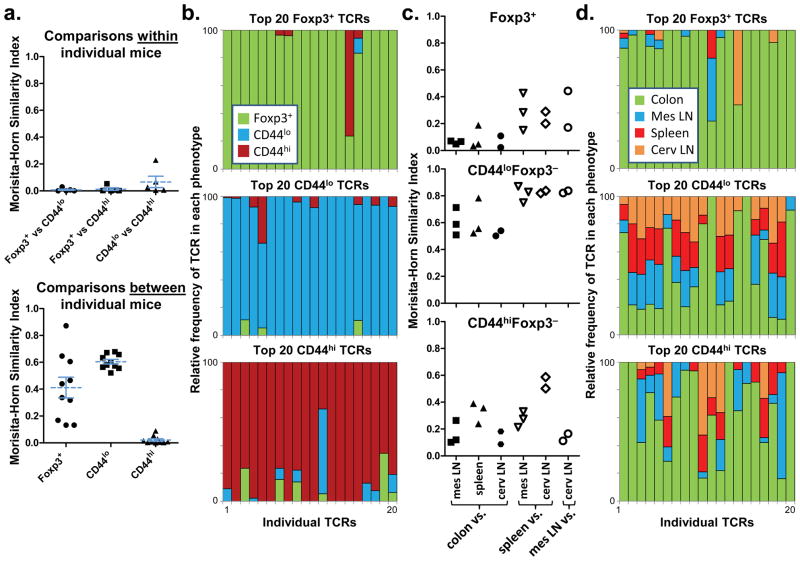
Figure 1. The colonic Treg TCR repertoire is unique.
(a) TCR usage between colonic T cell subsets. A total of 2,892 TRAV14 TCRα sequences from colonic naive, memory, and Treg cells of five individual mice were compared (Supplementary Fig. 2). Each symbol represents a Morisita-Horn similarity comparison between two different T cell subsets within each mouse (top), or a comparison of the same T cell subset between different mice (bottom). Bars represent mean ± S.E.M. (b) Analysis of individual TCRs. The relative distribution within all T cell subsets is shown for the 20 most common individual TCRs in each colonic T cell subset. For example, a TCR with equal percentage in the Foxp3+ and CD44hi subset would be shown as a half-green/half-orange bar. This analysis uses the pooled data set, which includes sequences from individual mice as well as 9,680 sequences from Expt. 1–3, each consisting of cells from 3–5 mice (Supplementary Fig. 2). Note that 1 TCR is found in both Foxp3+ (#15) and CD44hi (#3) plots, and 1 in both CD44lo (#5) and CD44hi (#12) plots; all others appear in only one plot. (c) Anatomic distribution of colonic TCRs. Morisita-Horn indices comparing the colon data to other locations(closed symbols), or between each of the other locations (open symbols), are shown. Abbreviations: mes, mesenteric; cerv, cervical. (d) Analysis of individual TCRs. The 20 most prevalent colon TCRs for each subset in the pooled data set are shown, and their presence at other locations represented in a manner analogous to (b).