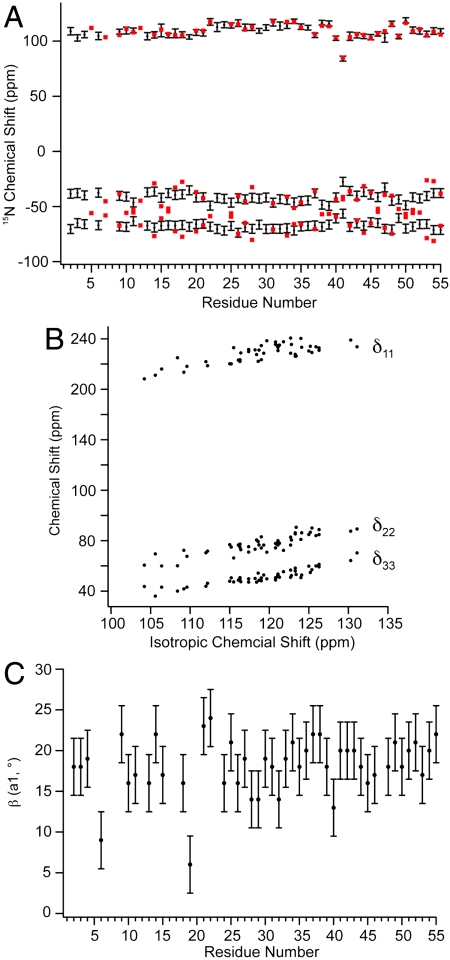
Fig. 5.
Amide chemical shift tensor analysis for protein GB1. (A) Principal elements of 15N tensor in the traceless representation compared to previously published slow-MAS data. Principal elements are presented in black with error bars corresponding to one standard deviation. Tensor values from previous work are presented in red. The rmsd between δ11 elements from both datasets is 1.6 ppm, and 6 ppm for δ22 and δ33 values corresponding to a deviation of η of 0.13. (B) Chemical shift tensor elements plotted against isotropic chemical shift. The correlation reveals that the changes in the isotropic chemical shift result from largely correlated shifts of all three principal elements. R2 for each element (δ11, δ22, δ33) are 0.82, 0.75, and 0.62, respectively. (C) The angle β (a1) as a function of residue number. The angle β defines the orientation of the δ11/δzz tensor element to the HN bond dipole.