Fig. 3.
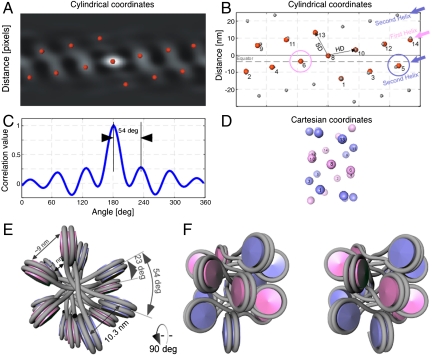
The fiber shows a two-start organization. (A) The autocorrelation function of the subtomogram average in cylindrical coordinates at a radius of 13 nm. The red spheres indicating the positions of the local maxima show a remarkable periodicity and are used to define the lattice points. (B) An idealized lattice, shown in gray spheres, was calculated based on the positions of the local maxima. In the idealized lattice the first helix is indicated by a pink arrow, and the second helix is indicated by a blue arrow. The positions of the nucleosomes visible in the real data are indexed by small numbers. Both helices are shown to intersect the equator at the positions indicated by the pink and the blue circle respectively. This shows that the fiber has a clear two-start geometry. (C) A plot through the autocorrelation function at the position of the basic helix shows that the subtomogram average has approximately 6.5-fold symmetry with a rotation per subunit of approximately54°. (D) Cartesian space depiction of the idealized lattice in B, facilitating indexing of the nucleosomes on the lattice. The numbering of the nucleosomes is identical to B. The two-start helix is visible in Cartesian coordinates. (E) Top view of the positions of the nucleosomes placed at the positions of the correlation peaks shown in B, with the linker DNA modeled in gray, which is assumed to cross the fiber-axis. The azimuthal rotation and center-to-center nucleosome distance are indicated. (F) Side view of the depiction in E (related by a 90° rotation) shown in crossed-eyes stereo view.