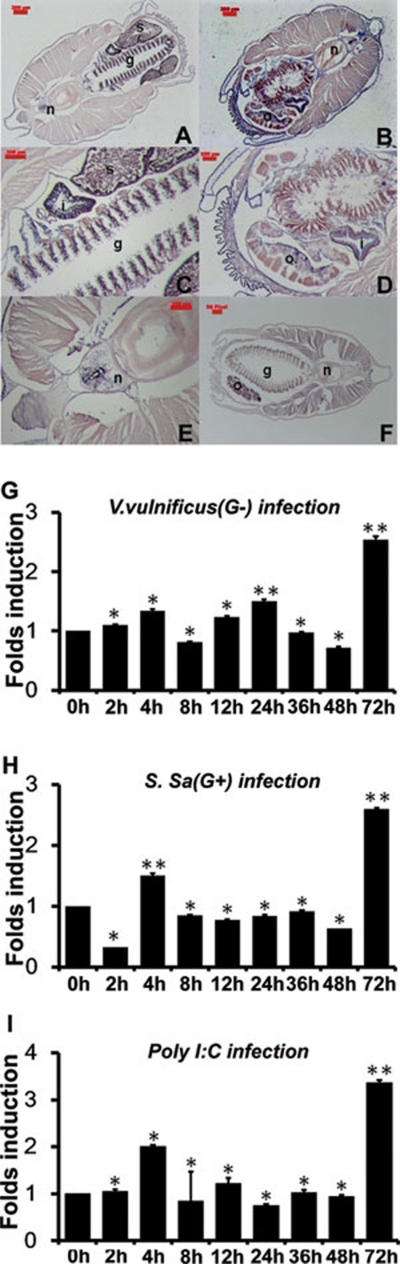
Figure 2.
The tissue distribution and expression pattern of bbtTICAM after challenge with bacteria and polyI:C. (A, B) Section in situ hybridization analysis of bbtTICAM anti-sense probe showed predominant expression in intestine, gill, and skin. bbtTICAM was not abundant in notochord. The magnification factor is 45. (C-E) Macroscopic view of the hybridization signals of bbtTICAM in gill, intestine, and notochord. The magnification factor is 150. A, C and E show the slice from a male adult, while B and D show the slice from a female adult. (F) Section in situ hybridization analysis of bbtTICAM using sense probe as negative control. n = notochord, i = intestine, g = gill, o = ovary, s = spermatozoa. The scale bars are shown in red. Blue indicates strong hybridization, and dark brown means a weak signal. (G-I) Quantitative real-time PCR (RT-PCR) analysis of the expression of bbtTICAM after challenge with gram-negative bacteria (G), gram-positive bacteria (H) and PolyI:C (I). Results were presented as fold induction of mRNA expression in triplicate from two parallel experiments, using 2-ΔΔCt method. Endogenous control for quantification was cytoplasmic β-actin. Values were considered to be significant at P < 0.05. Student's t-test (two-tailed distribution, two-sample unequal variance) was used for the calculation of all P-values. *P < 0.05 and **P < 0.01.