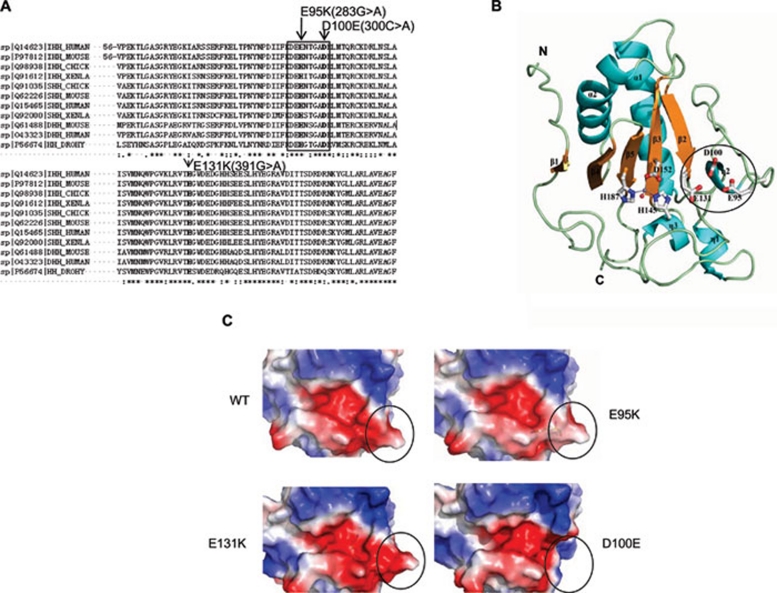
Figure 1.
Crystal structure and electrostatic potentials of human IhhN protein. (A) Comparison of the amino acid sequences of Drosophila, Xenla, chick, mouse, and human Hedgehog proteins. The mutations are shown in bold, and the eight-amino acid motif is indicated within the black pane. (B) Schematic representation of the overall structure of human IhhN. Secondary structures are labeled, with the α-helix and η-helix colored in cyan and the β-strand in yellow. The zinc ion is shown as the magenta dot, and the side chains of H145, D152, and H187 are shown in white with red oxygen atoms and blue nitrogen atoms. The zinc-bound water molecule is shown as a red dot. The N- and C-termini are individually labeled. The missense mutations at positions E95, D100, and E131, linked to BDA1, are labeled and shown as sticks. The eight-amino acid motif is indicated as a black circle. (C) Electrostatic potentials of human IhhN. Compared with WT, the E95K mutant changes the negatively charged area to positive. The E131K mutant exists in two alternative conformations, one of which is consistent with the WT, and the other forms a salt bridge between the carboxyl oxygen atom of E95 and the Nɛ atom of K131. The D100E mutant crystal structure showed high flexibility and local structure changes in the region comprising residues 93-98, indicated by a black circle. The electrostatic potentials of protein surfaces were scaled to the range of −5 (red) and 5 kT (blue), calculated using PyMOL 31 and APBS 32.