Abstract
We previously reported that XccR, a LuxR-type regulator of Xanthomonas campestris pv. campestris (Xcc), activates the downstream proline iminopeptidase virulence gene (pip) in response to certain host plant factor(s). In this report, we further show that the expression of the xccR gene was repressed in the culture medium by an NtrC-type response regulator, which we named XerR (XccR expression-related, repressor), and that this repression was relieved when the bacteria were grown in planta. Such a regulatory mechanism is reinforced by the observations that XerR directly bound to the xccR promoter in vitro, and that mutations at the phosphorylation-related residues of XerR resulted in the loss of its repressor function. Furthermore, the expression level of xccR increased even in XerR-overexpressing Xcc cells when they were vacuum infiltrated into cabbage plants. We also preliminarily characterized the host factor(s) involved in the above mentioned interactions between Xcc and the host plant, showing that a plant material(s) with molecular weight(s) less than 1 kDa abolished the binding of XerR to the xccR promoter, while the same material enhanced the binding of XccR to the luxXc box in the pip promoter. Taken together, our results implicate XerR in a new layer of the regulatory mechanism controlling the expression of the virulence-related xccR/pip locus and provide clues to the identification of plant signal molecules that interact with XerR and XccR to enhance the virulence of Xcc.
Keywords: Xcc, NtrC-like regulator, LuxR-like regulator, proline iminopeptidase, pathogen-host interaction, plant signal(s)
Introduction
In the past few decades it has become obvious that bacteria can display sophisticated group behaviors and form communities in their natural niches in response to constant changes in physical, chemical and biological environments 1, 2, 3. The regulation of gene expression mediated by signaling molecules and regulatory proteins in a bacterial population density-dependent manner is referred to as quorum sensing (QS). The first QS system in Gram-negative bacteria was observed in Vibrio fischeri, which contains a LuxR regulator and a cognate LuxI synthase responsible for producing autoinducer signal molecules N-acylhomoserine lactones (AHLs) 4, 5, 6. To date, QS-dependent functions have been studied in a wide variety of bacteria that control diverse bacterial processes, including virulence, sporulation, plasmid transfer, biosynthesis of antibiotics, as well as plant nodulation 7, 8, 9. It is now increasingly evident that QS is a complicated group behavior of bacteria for producing, sensing and responding to multifarious chemical signals to increase their chances of survival and propagation 7, 8. In other cases, QS-mediated communications are also involved in interactions between bacterial species and between bacteria and their hosts. For example, γ-amino butyric acid (GABA) produced by plant induces the expression of the attKLM operon in Agrobacterium tumefaciens, which causes the bacterium to destroy its own QS signal 10, while L-proline interferes with the import of GABA and antagonizes the degradation of bacterial QS signal, 3-oxo-octanoylhomoserine lactone 11.
A genomic survey of Proteobacteria showed that there are numerous bacteria that do not encode a cognate LuxI synthase for AHLs 12. As a result, the unpaired LuxR-like proteins designated as LuxR-family orphans or 'solos' have been studied 13, 14. LuxR solos such as ExpR of Sinorhizobium meliloti, BisR of Rhizobium leguminosarum pv. viciae and QscR of Pseuodomonas aeruginosa, respond to AHL signals produced by the bacteria themselves 15, 16, 17. In addition, SdiA in Salmonella, Escherichia and Klebsiella are able to bind and detect AHLs produced by other bacterial species 18. Interestingly, accumulating evidence from recent studies supports the idea that, apart from playing important roles in sensing AHL-like autoinducers, LuxR-like solos could potentially sense non-AHL signaling molecules as well 13, 14, 19.
As a special LuxR-like solo, XccR of the plant pathogen Xanthomonas campestris pv. campestris (Xcc) is required for activating the expression of the downstream proline iminopeptidase gene (pip) through binding to the luxXc box in the pip promoter, and this activation is enhanced by plant host factors 20. The xccR/pip locus is different from the classical luxR/luxI system in that pip is a virulence-related gene, rather than a gene for producing AHL signals. The xccR/pip-like locus has been found in several other bacteria, such as S. meliloti, Rhodospirillum rubrum, R. leguminosarum and P. syringae 20. More particularly, the oryR/pip locus of Xanthomonas oryzae pv. oryzae (Xoo) behaves very much like the xccR/pip locus. In addition, the solubility of OryR is enhanced by a rice extract with molecular weights less than 1 kDa 21. OryR also positively regulates the expression of a cell wall-degrading cellobiosidase gene for optimal pathogenicity 22.
In this study, we explored the bacterial upstream factor(s) and the host plant signals regulating the expression of the xccR/pip locus. By screening a genome-scale Tn5-insertion library of an Xcc strain harboring an xccR promoter-gusA fusion, we identified an NtrC-type transcriptional regulator XC_3760 (named XerR, XccR expression-related, repressor) as a repressor of the xccR/pip locus. NtrC-type proteins have been recognized as enhancer-binding proteins in phosphorylated forms; they are involved in nitrogen assimilation, biofilm formation, bioluminescence and QS regulatory system, and thus their functions are expected to be pleiotropic 23, 24, 25, 26. Furthermore, we showed that the repressor function of XerR was relieved in the presence of the host plant extract with molecular weights less than 1 kDa, and that the same plant extract enhanced the binding of XccR to the pip promoter sequence. Our results expand the regulatory machinery controlling the expression of the pathogenicity-related xccR/pip locus and provide new insights into how Xcc senses host signals to regulate its infectivity.
Results
Genetic screening of xccR expression reveals a repressor, XerR
To identify factors that regulate the expression of xccR, which directs the expression of the virulence gene pip in Xcc, we designed an antibiotic-coupled transposon screen. The chromosomal xccR promoter (xccR-P)/gusA fusion strain (Xcc 8177) was mutated with the EZ-Tn5 transposon that contains the dihydrofolate reductase (DHFR) gene for conferring trimethoprim resistance. From 20 000 transposon-insertion mutants, we selected one that pointed to a possible repressor of xccR expression. Analysis of the flanking sequences of the mutated gene indicated the gene was XC_3760 encoding a transcriptional regulator of the NtrC family, which we designated as xerR in this paper.
XerR is a putative 433 amino acid protein with a predicted molecular weight of 48.2 kDa and belongs to the two-component signal transduction system (TCSTS) response regulator (RR) NtrC family 27. BLAST search against the databases revealed that XerR is highly conserved in all Xanthomonas species and shares significant sequence similarities with NtrC family proteins from Bordetella bronchiseptica (61%), Ralstonia solanacearum (60%) and Burkholderia xenovorans (58%; NCBI Blast: http://www.ncbi.nlm.nih.gov/BLAST/). Analyses with Pfam indicated that XerR contains an N-terminal receiver domain that has the conserved Asp site for phosphorylation, a central ATP-binding AAA+ domain that hydrolyzes ATP to generate energy, and a C-terminal domain containing a helix-turn-helix motif for DNA binding 23. Multiple sequence alignments between XerR and CheY in Escherichia coli 28, NtrC in Salmonella typhimurium 29 and LuxO in Vibrio harveyi 25, revealed the highly conserved residues Asp-17, Asp-60 and Phe-106 in the receiver motif (Figure 1A). These residues were shown to be critical for the functioning of the phosphorylated protein, and Asp-60 was proposed to be the phosphorylated site. Although the histidine kinase and its cognate RR are usually linked in one operon 30, a search of the genome did not suggest that the Xcc chromosome encodes a cognate TCSTS sensor protein in close vicinity of the xerR sequence. It was the overall structure and location of xerR that prompted us to study its biological functions.
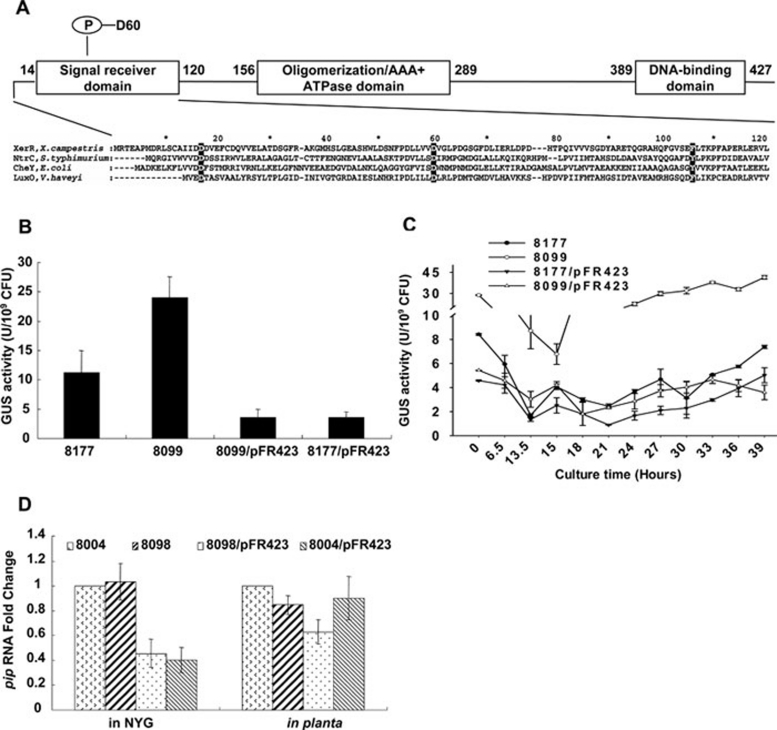
Figure 1.
XerR is required for repression of xccR and pip transcription in medium. (A) The domain organization of XerR and the sequence of receiver domain. Three putative modular components of XerR are shown in the diagram. Multiple amino acid sequence alignments between XerR and NtrC in S. typhimurium, CheY in E. coli and LuxO in V. harveyi are shown at the bottom of the diagram. The residues altered by site-directed mutagenesis are shaded in black, and the putative phosphorylation site (Asp-60) is marked. (B) GUS expression levels in different Xcc strains were assayed by enzymatic activities. xerR in-frame deletion mutant Xcc 8099 increased the GUS activity compared to that of Xcc 8177. Xcc 8099 and Xcc 8177 carrying the xerR gene in pHM1 plasmid (Xcc 8099/pFR423 and Xcc 8177/pFR423) exhibited reduced GUS activities. All the strains were harvested at OD600 of 2.0 in NYG medium. Relative GUS activity units were defined as nM 4-methylumbelliferyl/min/109 cells. The means and standard deviations were calculated from the data derived from at least nine independent experiments. (C) Expression of xccR-P/gusA in Xcc 8099 was density dependent when grown in NYG medium. GUS activities of different strains were assayed at different time points. The mean and standard deviation were calculated from the data derived from three independent experiments. (D) Expression levels of pip in different Xcc strains in medium and in planta. Relative transcriptional levels of pip were quantified by real-time RT-PCR. In NYG medium, RNA were extracted from the cultured strains at a cell density of OD600 = 1.5−2.0. In planta, RNA was isolated from vacuum-infiltrated cabbage leaves 30 h post infiltration. Measurements were normalized by the wild-type values and fold differences were plotted. Each sample was assayed in triplicate.
To verify the role of XerR in xccR repression, an xerR non-polar markerless deletion was introduced into the chromosome of Xcc 8004 to generate the xerR mutant Xcc 8098, and then an xccR-P/gusA fusion was inserted into Xcc 8098 to create Xcc 8099. β-Glucuronidase (GUS) activities in Xcc 8099, its complementation strain Xcc 8099/pFR423, in which pFR423 carries xerR driven by the lacZ promoter, and the XerR-overexpression strain Xcc 8177/pFR423 were examined and compared with that in Xcc 8177 at mid-exponential phases of bacterial growth (Figure 1B). The results showed that GUS activity in Xcc 8099 was increased 2.14-fold relative to Xcc 8177, whereas the GUS activities in both complementation and overexpression strains were reduced to 30% of that of Xcc 8177. Although GUS activities in Xcc 8177 did not display a typical QS behavior and stayed at low levels as those in Xcc 8099/pFR423 and Xcc 8177/pFR423, GUS levels in Xcc 8099 increased along with the cell growth (Figure 1C). These data suggest that de-repressed expression of xccR occurs in a density-dependent manner.
We previously showed that under medium culture conditions, overexpression of XccR significantly enhanced the expression of the downstream pip gene, while in wild-type Xcc 8004 pip expression remained very low throughout the bacterial growth phases. Here we tested whether the XerR protein has an indirect effect on pip transcription. By using real-time reverse transcription (RT)-PCR, we found that in XerR-overexpression strains Xcc 8098/pFR423 and Xcc 8004/pFR423, the level of the pip transcript decreased significantly by 55% and 60%, respectively (Figure 1D). One possibility is that XerR also potentially repressed the expression of pip. However, the pip RNA levels in xerR-deleted Xcc 8098 showed little increase compared with that in Xcc 8004. Furthermore, when Xcc 8004 and Xcc 8098 were grown in NYG medium, no XccR protein was detected with anti-XccR antibodies (data not shown). The result is similar to OryR protein, a homolog of XccR in Xoo, which was also not detectable by western blot analysis when bacteria were grown in minimal M9 medium 21. These results indicate that the increased expression of xccR by xerR mutation was not sufficient to provide enough stable XccR protein to alter the pip RNA level.
XerR acts as a repressor by binding to the xccR promoter
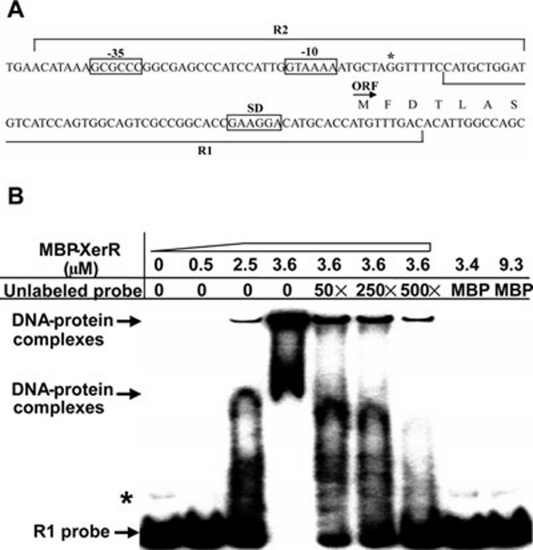
Usually two-component RR NtrC family proteins act on phosphorylation as enhancer-binding proteins via interaction with σ54. However, analysis of the xccR promoter sequence did not reveal a highly conserved σ54-recognition sequence GG-N10-GC 31. We thus explored the possibility that XerR directly interacts with the xccR promoter. Electrophoretic mobility shift assays (EMSAs) were performed using purified XerR protein tagged with an N-terminal MBP and DNA sequences upstream of the xccR coding region as probes, which spanned −50 to +9 (R1) and −99 to −40 (R2), respectively, relative to the translational start site (Figure 2A). Addition of MBP-XerR to the reaction mixtures caused a shift in the mobility of R1 fragment (Figure 2B) and R2 fragment (Figure 4C), but addition of pure MBP did not. The shifted bands could be competed by 50-fold excess of the unlabeled probes, indicating a specific binding of XerR to the xccR promoter. The binding affinity of XerR affirms its ability to repress the xccR transcription, and as a repressor it likely prevents RNA polymerase from binding to transcriptional sites and ensures that the gene is turned off in an efficient and specific manner 32, 33.
Figure 2.
EMSA shows that XerR binds to the upstream region of the xccR gene directly. (A) Schematic of the upstream region of xccR gene according to a promoter prediction program NNPP version 2.2 (1999). The putative −35/−10 and SD (Shine-Dalgarno sequence) elements are boxed, and an asterisk denotes the xccR transcriptional start site. The locations of two different probes that have a 10-bp overlap are denoted by lines. (B) EMSA assay of R1 probe with purified MBP-XerR. Isotope-labeled probe (8 fmol) was incubated for 30 min with indicated concentrations of protein (in μM) at room temperature. The shifted bands could be competed by excess of the unlabeled probe. The folds of unlabeled probe were indicated above. The migrated DNA-protein complexes and free probe R1 are indicated by arrows, and the bands marked with an asterisk indicate a possible higher structure of R1 probe formed during annealing step.
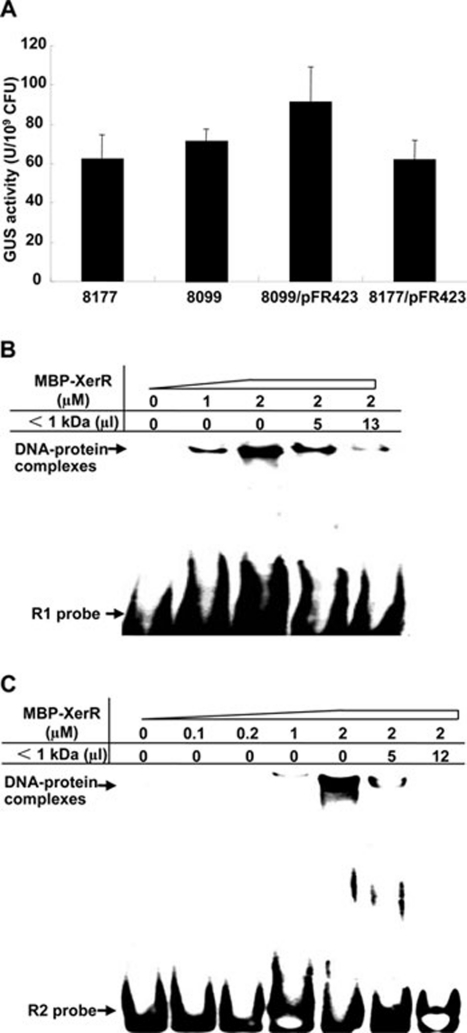
Figure 4.
XerR relieves its inhibition on xccR expression in planta. (A) In planta cultivation did not significantly increase the GUS activity from Xcc 8099, while it had the opposite influence on that of Xcc 8099/pFR423 and Xcc 8177/pFR423. The bacteria were recovered from vacuum-infiltrated cabbage leaves 30 h post infiltration, and GUS activities were assayed. Data and standard deviation represented the mean of three independent measurements. (B and C) Plant signal(s) alleviated the binding activity of XerR protein to the xccR upstream regulatory sequence. EMSA assays with biotin-labeled probe were performed by MBP-XerR with plant extracts (< 1 kDa) at two dilutions.
Phosphorylation-related residues of XerR are essential for its repressor function
In vivo and in vitro experiments indicated that XerR can efficiently repress the expression of xccR. To characterize the repressor function-related motifs, we first tested whether the N-terminal part or the C-terminal part of XerR is critical in regulating transcription of xccR. We constructed an xerR RR domain (N-terminal amino acids 14-120) deletion mutant (xerR ΔRR) and an HTH domain (C-terminal amino acids 385-433) deletion mutant (xerR ΔHTH), and assayed GUS activities under the control of the xccR promoter in these two xerR deletion strains. The results showed that the GUS activities produced by xerR ΔRR and xerR ΔHTH were 3.06 times and 2.97 times, respectively, of that of Xcc 8177 in NYG medium (Figure 3A), indicating that removal of the receiver or DNA-binding domain results in an inactive XerR protein and thus de-represses the xccR transcription.
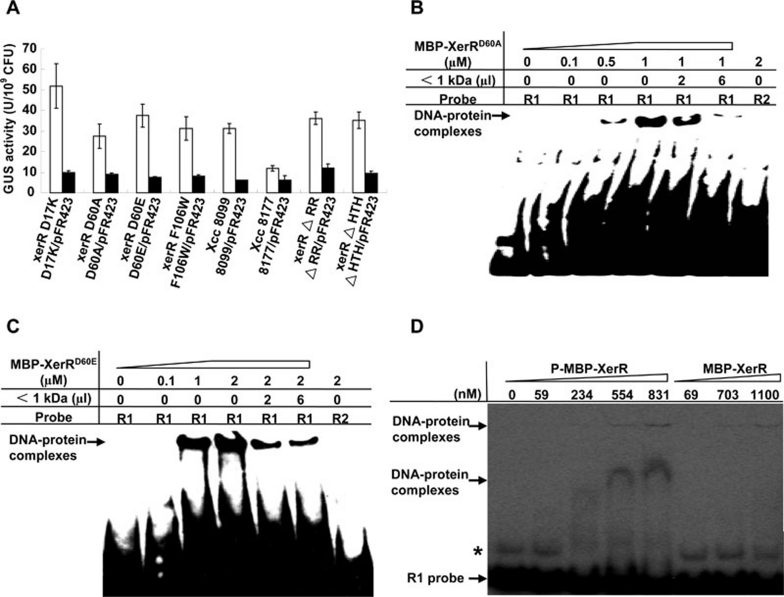
Figure 3.
Phosphorylation-related residues of XerR are essential for its repressor function. (A) The conserved phosphorylation-related residues and the regulatory domains of XerR were indispensable for regulation of xccR expression in vivo. xerR ΔRR, xerR ΔHTH and different site-directed mutants exhibited increased GUS activities when grown in NYG medium. Plasmid-containing (pFR423) strains of different mutants reduced the GUS activities compared with that of Xcc 8177. Bacteria cultured in NYG medium were assayed at an OD600 of 2.0. The experiments were repeated eight times with similar results. (B and C) EMSA assays of MBP-XerRD60A and MBP-XerRD60E with biotin-labeled R1 probe and plant extract. The two proteins presented the same binding characteristics to R1 probe, in which the plant extract of molecular weights < 1 kDa released the protein and DNA interactions. In the diagram, the concentration of purified protein and the volumes of plant signal(s) are indicated. (D) EMSA binding of phosphorylated and unphosphorylated MBP-XerR to the R1 probe. MBP-XerR was phosphorylated in vitro with acetyl phosphate and the R1 probe was end-labeled with 32P at its 5′ termini. The bands marked with an asterisk indicate a possible higher structure of R1 probe formed during annealing step.
Next, we investigated the roles of the phosphorylation-related residues Asp-17, Asp-60 and Phe-106 in the repressor function of XerR. We constructed four xerR site-directed mutants on the Xcc 8177 background (xerR D17K, xerR D60A, xerR D60E and xerR F106W) and assayed their GUS activities (Figure 3A). Similar to the xerR null mutant Xcc 8099, each site-directed mutant showed a considerably higher GUS level than Xcc 8177, suggesting that the canonical phosphorylation-related residues are required for XerR function in vivo. Furthermore, the low-copy plasmid carrying wild-type xerR (pFR423) was able to restore the XerR repressor activity in trans in all of the xerR deletion and site-directed mutants (Figure 3A).
In addition, we analyzed whether the Asp-60-mutated proteins MBP-XerRD60A and MBP-XerRD60E can still bind the xccR promoter sequences, since Asp-60 was proposed to be the phosphorylation site by Pfam alignment. We found that although both mutated proteins could bind to R1, they lost the ability to bind R2 even at higher protein concentrations (Figure 3B and 3C), suggesting that XerRD60A and XerRD60E have altered DNA binding properties and thus cannot repress the xccR promoter.
On the other hand, we found that phosphorylation of XerR enhanced the binding to R1. As shown in Figure 3D, in vitro phosphorylated XerR (P-MBP-XerR) exhibited an affinity to bind R1 in EMSA at a concentration of 554 nM, which is lower than that needed for unphosphorylated MBP-XerR protein. Under an equivalent condition, we did not observe the band-shift at 1.1 μM for unphosphorylated MBP-XerR protein.
Taken together, the above results indicate that phosphorylation of XerR is essential for its repressor function, reminiscent of the intrinsic property of an NtrC family protein.
Inhibition of xccR expression by XerR is relieved in planta
We previously reported that the expression of xccR and pip was induced when the Xcc cells grew in the host cabbage 20. In this report, we showed that XerR inhibited the expression of xccR and pip in culture medium. To see if the XerR-mediated inhibition is affected in planta, we quantified and compared the xccR expression levels in planta (Figure 4A) and in NYG medium (Figure 1B) in different Xcc strains. Overexpression of XerR in Xcc 8099/pFR423 and Xcc 8177/pFR423 greatly reduced the xccR promoter-directed GUS activities in NYG medium compared with that of Xcc 8177, as shown in Figure 1B. However, the GUS activities in the XerR-overexpressing strains were not reduced or even increased relative to that of Xcc 8177 when the bacteria grew in planta (Figure 4A). Furthermore, the Xcc 8099 strain had almost equivalent GUS activity as Xcc 8177 (Figure 4A), suggesting that the repression action of XerR on xccR expression might be relieved in planta.
As pip expression is controlled by XccR, we expected the inhibition of expression of pip by XerR in culture medium would also be relieved in planta. This was actually the case. As seen in Figure 1D, the significantly reduced pip transcript levels in XerR-overexpressing strains Xcc 8098/pFR423 and Xcc 8004/pFR423 in culture medium were restored to 63% and 90% of that of wild-type Xcc 8004, respectively, when the bacteria grew in planta. We reasoned that the increased expression of xccR and pip was not a result of reduced transcription of xerR gene in host plant, because the GUS activity of xerR-P/gusA in planta was 2.56-fold higher than that in medium alone (data not shown). In addition, the expression of XC_3756, another gene that is directly regulated by XerR via binding to the σ54 cis-element in its promoter, was enhanced threefold in planta compared with that in medium (data not shown).
To examine whether the observed de-repression of xccR expression was caused by plant factors that affected the binding of XerR to the xccR regulatory sequences, we performed EMSA in the presence of a low-molecular-weight (< 1 kDa) cabbage extract. As shown in Figure 4B and 4C, the presence of the plant extract disrupted the binding of XerR to the xccR promoter probes R1 and R2 in a dose-dependent manner. We infer from this result that some small molecules present in this cabbage extract are responsible for limiting the XerR binding to DNA. This interfering effect was DNA sequence specific, as the same amount of the cabbage extract did not cause dissociation of XerR from the promoter of another downstream gene XC_3756 (data not shown). We further showed that the same extract also abrogated the ability of the mutant proteins MBP-XerRD60A and MBP-XerRD60E to retard the migration of the DNA probe R1 (Figure 3B and 3C), indicating that the two amino acid substitutions for Asp-60 of XerR do not alter the interaction between XerR and the plant signal(s).
Cabbage extract enhances the binding of XccR to the pip promoter
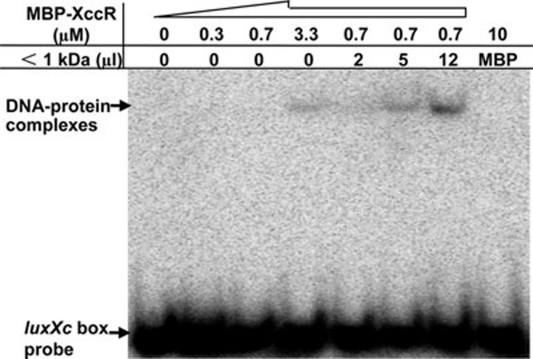
We previously reported that in a super-shift assay, a cabbage ethanol extract enhanced the binding of XccR to the pip promoter 20. In addition, it was reported that an unknown rice signal molecule present in the < 1 kDa fraction of an Xoo-infected rice extract increased the solubility of OryR 21. We thus wanted to see if the < 1 kDa cabbage extract, which abolished the binding of XerR to the xccR promoter, could affect the binding of XccR to the luxXc box of the pip promoter. We found that the < 1 kDa extract indeed stimulated the binding of MBP-XccR to the luxXc box sequence at an MBP-XccR concentration of 0.7 μM at which no protein-DNA binding occurred without the plant extract (Figure 5). Moreover, the formed protein-DNA complex was significantly intensified with an increase in the concentration of the plant extract (Figure 5). The above results indicate that the same cabbage extract shows different effects on the formation of the XerR/xccR promoter complex and the XccR/pip promoter complex. Although we cannot conclude that XerR and XccR interact with the same compound in the cabbage extract, our results portray a subtle regulatory pattern in which Xcc recruits plant signal(s) to sequester XerR from its binding sequence, yet on the other hand, to stimulate the XccR binding to the pip promoter for infectivity
Figure 5.
EMSA binding of MBP-XccR protein to the luxXc box of the pip promoter. The band of XccR and DNA complex was intensified by adding different volumes of plant extracts in the EMSA assay. The migrated DNA-protein complexes and isotope-labeled luxXc box probe are indicated by arrows.
Discussion
In Gram-negative bacteria, LuxR/LuxI is the most well-defined regulatory system that modulates gene expression related to QS. This system can monitor the concentration of AHL-like small molecules in the environment and control downstream gene expression or cell behavior 34, 35. In Xanthomonas, the solo (orphan)LuxR homologs, including XccR from Xcc and OryR from Xoo, can sense chemical signals derived from host plants and take part in bacterial pathogenesis by regulating the expression of virulence factors 20, 21, 22. These studies strongly suggested an interesting phenomenon that an inter-kingdom communication exists between phytopathogens and their host plants. However, how bacteria sense signals from plant and the nature of the plant signal(s) remain unclear.
In this study, an NtrC family RR was identified by genome-scale screening with a transposon insertional mutant library. By measurement of GUS activity, XerR was confirmed to be a negative regulator of xccR expression (Figure 1B). XerR can bind R1 and R2 regions of xccR upstream sequence (Figures 2B and 4C). It was shown that mutations of the conserved phosphorylation-related sites on XerR resulted in upregulation of xccR expression (Figure 3A) and that in vitro phosphorylated XerR showed enhanced affinity to the R1 probe (Figure 3D), both results suggesting that protein phosphorylation is required for the repression function of XerR. In addition, GUS assays in planta (Figure 4A) as well as EMSA experiments (Figure 4B and 4C) showed that the binding of XerR to the xccR upstream DNA sequence was substantially inhibited in the presence of the plant extract, suggesting that a plant signal(s) modulates the xerR/xccR/pip regulatory cascade.
By secondary protein structure prediction (Figure 1A), XerR was found to be a typical NtrC-family RR of the bacterial TCSTS. It contains an N-terminal CheY-like receiver domain, a C-terminal HTH domain, and a central σ54 interaction domain responsible for the initiation of an open transcriptional complex 36. In prototypical TCSTS, a histidine kinase (HK) sensor can monitor specific environmental stimuli. After autophosphorylation on a conserved histidine residue, the dimeric HK transfers the phosphoryl group onto the conserved asparagic acid residue of the cognate RR, and the latter will regulate downstream gene expression, usually acting as a transcription factor 37, 38. As mentioned above, site-directed mutations, which changed the three critical sites (Asp-17, Asp-60 and Phe-106) related to protein phosphorylation, nearly abolished the repressor activity of XerR (Figure 3A). In addition, XerR can receive the phosphorylation signal in vitro (Figure 3D). These results suggest that under the culture conditions, an unidentified HK can phosphorylate XerR specifically and cause it to bind to the xccR promoter with high affinity. Transcriptional repression of xccR may result from the direct competition of phosphorylated-XerR and RNA polymerase on its promoter or the blocking of mRNA elongation, as the probes that we used spanned from −99 to +9 bp relative to the xccR translational start site. The repression mechanism of XerR remains to be fully elucidated. Intriguingly, XerRD60A and XerRD60E abolished the binding ability to R2 (Figure 3B and 3C), whereas the E. coli NtrCD60A and NtrCD60E proteins bound to the cognate enhancer sequence normally 39, 40.
In addition, our results also showed that the repressor function of XerR was relieved when Xcc was grown within the host plant (Figure 4A). In TCSTS, the central mechanism is the balance of phosphorylated and dephosphorylated RR 41, 42. Since the majority of HKs also have phosphatase activity to dephosphorylate RR 43, 44, 45, we speculated that the cognate HK of XerR in planta may exhibit phosphatase activity, rather than acting as a kinase, which would result in the dephosphorylation of XerR and thus decrease its affinity in binding the xccR promoter. Because there is no HK gene located in the vicinity of xerR, it is extremely difficult to identify its cognate HK only by bioinformatic analysis. Currently we have identified a candidate HK gene that also exhibits repressor function in regulating xccR transcription (unpublished data). Further investigation will clarify whether it is the cognate HK of XerR and how it modulates the phosphorylation state of downstream RR when Xcc survives in different ecological niches.
Besides phosphorylation-regulated xccR expression via classical TCSTS, our experiments also suggest that the activity of XerR is affected by chemical signal(s) from host plants. As shown in Figure 4B and 4C, when the plant extract was present in EMSA, the binding between XerR and the xccR promoter was disrupted in a dose-dependent manner. We found that the expression of xerR was increased in planta, and XerR-mediated upregulation of the transcription level of XC_3756 was not affected when Xcc was grown in the host plant, implicating that the unidentified plant chemical(s) has a specific influence on the binding between XerR and the xccR promoter. Inter-kingdom communications involved in de-repressing the expression of a bacterial gene by plant signal(s) also occur in other bacterial-eukaryotic systems. For instance, the repression of pectinase genes by the transcriptional repressor KdgR in Erwinia carotovora was abolished in the presence of plant cell wall breakdown products, and agrocinopines de-repressed the AccR binding activity to the arc operon in A. tumefaciens 46, 47, 48. Intriguingly, the same plant extract improved the binding of XccR to the luxXc box of the pip promoter, indicating that XerR and XccR may simultaneously interplay with plant signal(s) in regulation of the xccR/pip locus, imposing a strict control on the expression of a virulence gene.
It was shown that host plant signal(s) with molecular weights less than 1 kDa inhibited the repression function of XerR in a concentration-dependent manner (Figure 4B and 4C). Although most of the small signaling molecules to date were extracted by organic solvents, including furanones, flavonoid, riboflavin and its derivative lumichrome 49, 50, 51, 52, our cabbage extract was water based. We propose that the signal(s) may be peptides, amino acid or its derivatives, monosaccharides, oligosaccharides, aminosugars, aminoglycosides or acid and alkali compounds. It has been reported that two free amino acids, homoserine and asparagine, act as host signals inducing the pelD expression of Nectria haematococca in pea seedlings 53.
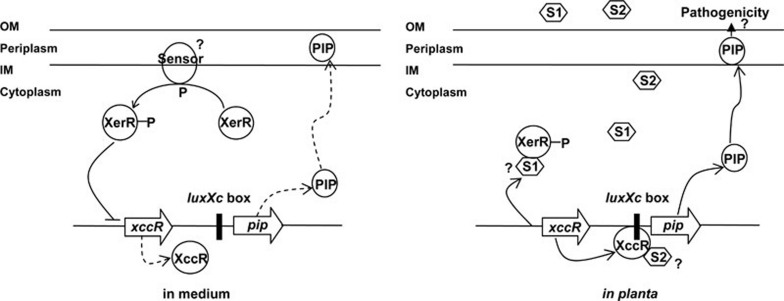
In brief, in the interaction between Xcc and the host plant, Xcc builds up a sophisticated mechanism in the xerR/xccR/pip pathway. In this pathway plant signal(s) either activates XccR to positively regulate pip transcription or relieves the inhibition of XerR on xccR expression. As illustrated in Figure 6, we propose a model that the transcriptional inhibitory role of XerR is regulated by an unknown HK, and XerR then blocks the transcription of xccR by directly binding to the xccR upstream sequence during growth in the medium. When Xcc grows in the host plant, the conformational changes of XerR and XccR induced by the plant small molecule(s) lead to the release of XerR from the xccR promoter and the increase in the binding of XccR to the luxXc box. Consequently, the expression of the pip gene, which plays a crucial role in bacterial pathogenesis, is fine-tuned in the host plant.
Figure 6.
A model for expression regulation of xccR/pip locus by XerR. When Xcc is grown in NYG medium, XerR represses the expression of xccR and pip. The activity of XerR is dependent on the phosphorylation by an unknown two-component signaling transduction system. After entering the host plant, XerR relieves its inhibition to xccR expression in response to specific plant small molecule(s). The released XccR binds to luxXc box in the presence of the same or different plant signal(s) to induce the transcription of the pip gene for bacterial virulence. S1 and S2 denote the possible signal(s) from the host. OM and IM refer to the outside and inside membrane, respectively.
Materials and Methods
Strains and reagents
The wild-type Xcc strain 8004 and Xcc 8177 harboring a chromosomal xccR-P/gusA fusion in Xcc 8004 were described previously 20, 54. Xcc strains were routinely cultivated at 28 °C in NYG medium, whereas E. coli strains were grown aerobically at 37 °C in LB medium. Antibiotics were added at the following concentrations: for Xcc, rifampicin (100 μg/ml), spectinomycin (150 μg/ml) and kanamycin (100 μg/ml); for E. coli, spectinomycin (150 μg/ml), kanamycin (50 μg/ml) and ampicillin (100 μg/ml). The reagents 5-bromo-4-chloro-3-indolyl-β-𝒟-glucuronic acid (X-Gluc), 4-methylumbelliferyl-β-𝒟-glucuronide (MUG), 4-methyl-umberlliferone and lithium potassium acetyl phosphate were purchased from Sigma. RNase-free DNase, M-MLV Reverse Transcriptase and Random Primers were from Promega and the Lightshift Chemiluminescent EMSA kit was from Pierce.
Creation of Xcc mutants and preparation of in trans expression constructs
Deletion strains were generated with the suicide vector pK18mobSacB 55 by a long-flanking homology procedure and two-step recombination 56. All DNA manipulations were performed according to standard procedures. Plasmid DNA was transferred to E. coli by heat shock and to Xcc strains by electroporation. Unless otherwise specified, the corresponding gene fragments were PCR amplified and first cloned into pEASY-T1 Vector (TransGen Biotech) for sequence verification. After digestion with appropriate enzymes, these fragments were cloned into corresponding vectors to generate the constructs used in this study.
To construct Xcc 8098, two ∼500-bp sequences upstream and downstream of the xerR reading frame were amplified by PCR. The in-frame deletion resulted in removal of the codons for amino acid residues 8 to 428. After digestion with appropriate enzymes, a pK18xerR clone was created by cloning two recovered fragments into pK18mobSacB simultaneously. The pK18xerR plasmid conferring kanamycin resistance (KanR) and sucrose sensitivity (SucS) from white colonies was verified by restriction digestions or by sequencing, and then transferred to Xcc. Allelic replacement was achieved by sequential selections on kanamycin and 10% sucrose to create Xcc 8098. Positive transformants of Xcc 8098 were confirmed by PCR and sequencing. To introduce xccR-P/gusA into Xcc 8098, a 3.2-Kb DNA fragment carrying the xccR promoter and the gusA gene was cloned into pK18mob vector to generate plasmid pFR435. The suicide vector pFR435 was integrated into the chromosome of Xcc 8098 by homologs recombination via a 542-bp sequence of the xccR promoter. The resultant strain was termed Xcc 8099. PCR was used to identify positive transformants, and the PCR products were sequenced. The same procedures were applied in generation of the other two deletion mutants: xerR ΔRR and xerR ΔHTH.
To perform in trans expression analyses, the entire coding sequence of the xerR gene was PCR-amplified and constructed into pHM1, a broad host range expressing plasmid 57. The generated clone was sequence verified and named pFR423. pFR423 was then electrotransformed into xerR null mutant 8098, 8099 and Xcc 8004, resulting in the complemented and overexpressed strains Xcc 8098/pFR423, Xcc 8099/pFR423 and Xcc 8004/pFR423, respectively. The resultant transformants were selected on NYG medium supplemented with rifampicin and spectinomycin. Plasmid derivatives harboring the correct inserts in Xcc strains were extracted and verified by restriction digestions.
Site-directed mutagenesis
Three conserved amino acid residues of XerR (Asp-17, Asp-60 and Phe-106, in the RR domain) predicted to be involved in phosphorylation were identified via the sequence alignment with the homologs: NtrC (NCBI accession number X85104), CheY (M13463) and LuxO (L26221). A 753-bp fragment containing part of the xerR gene and its flanking sequence was PCR-amplified and cloned into pEASY-T1, resulting in pEASY-T753. Three conserved residues were changed to lysine (D17K), alanine (D60A), glutamate (D60E) and tryptophan (F106W) by site-directed mutagenesis (Easy Mutagenesis Systems, TransGen Biotech) using the pEASY-T753 vector as the template. The four resultant 753-bp mutant fragments were separately inserted via SpeI and PvuI sites into pK18xerR-28, a plasmid containing the xerR gene and its up- and down-stream flanking sequences in pK18mobSacB. By homologs recombinations, the mutation-containing plasmids pK18xerR-D17K, pK18xerR-D60A, pK18xerR-D60E and pK18xerR-F106W were individually incorporated into Xcc 8098. Positive clones were verified by PCR and DNA sequencing. The same homologs recombination procedures were used to insert the xccR-P/gusA cassette into different mutants to create xerR D17K, xerR D60A, xerR D60E and xerR F106W.
GUS assay
GUS assays were used to examine the expression of the xccR-P/gusA fusion in different Xcc strains. GUS activity of the bacteria grown in medium and in planta was measured by the fluorometric method using MUG as a substrate essentially as described in Zhang et al. 20.
RNA extraction and RT-PCR analysis
Bacterial cells at OD600 of 1.5 to 2.0 were harvested by centrifugation at 4 °C for 2 min at 12 000× g. Total bacterial RNA was isolated using TRIzol reagent (Invitrogen) following the protocol provided by the manufacturer. RNase-free DNase I was used to treat the RNA samples. RT was performed using M-MLV Reverse Transcriptase with random hexadeoxynucleotides as primers. Typically, 25 ng of cDNAs was used for each PCR reaction in a 25 μl mixture. The 16S rRNA was served as an RT-PCR internal reference.
To assay the level of the pip transcript in Xcc strains grown in planta, Xcc cells were vacuum infiltrated into cabbage seedlings as described by Zhang et al. 20. Plant leaves harvested at 30 h post infiltration were homogenized in liquid nitrogen, and RNA isolation and RT-PCR were performed as described above. RNA from un-infiltrated leaves was used as a control.
Protein expression and purification
Prokaryotic expression plasmids pMX766 (pMal-p2X(lac-P/xerR)), pMX767 (pMal-p2X(lac-P/xerR-D60A)), pMX768 (pMal-p2X(lac-P/xerR-D60E)), pMX769 (pMal-p2X(lac-P/xccR)) were transformed into E. coli TB1. Two milliliters of the overnight culture were inoculated into 200 ml LB broth plus 2% glucose and ampicillin. After the cells were grown at 37 °C to an OD600 of 0.5, MBP-fusion proteins were induced by addition of IPTG to a final concentration of 0.3 mM and cell growth was continued overnight at 16 °C with a gentle shaking at 180 r.p.m. The cells were harvested, and the soluble MBP-tagged proteins were purified by affinity chromatography with amylose resin (BioLabs). Briefly, each cell pellet was resuspended in 10 ml of the column buffer (20 mM Tris-HCl, pH 8.0; 200 mM NaCl; 1 mM EDTA) plus 1 mM PMSF, incubated on ice for 20 min, sonicated, centrifuged and the supernatant was added to 2 ml of amylose resin slurry. After washing six times with the column buffer, the proteins were eluted using 10 ml of the column buffer plus 10 mM maltose three times. The purified protein samples were combined, and the solvent was changed to a buffer containing 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 1 mM DTT, 5% glycerol. Finally, the proteins were concentrated to approximately 1-10 mg/ml using Amicon YM-10 column (Millipore) and filtered through an Ultrafree-MC (0.45 μm) spin filter (Millipore) before aliquoting for storage at −80 °C. Protein concentrations were measured using the Bio-Rad Protein Assay reagent with BSA as a standard. About 5 μg of each protein sample was analyzed by 8% SDS-PAGE to verify molecular weight and purity.
Electrophoretic mobility shift assay
MBP-XerR and MBP-XccR fusion proteins were purified through amylose columns as described above. Four 59-nt single-stranded DNA oligonucleotides containing putative XerR-binding sequences upstream of the xccR coding region and two 46-nt pip promoter sequences with or without biotin labeling were synthesized by Invitrogen. DNA duplexes required for EMSA were annealed by mixing equal amounts of single-stranded oligos and incubating the mixture for 10 min at 93 °C in annealing buffer (10 mM Tris-HCl, pH 7.5; 1 mM EDTA; 100 mM NaCl). After slowly cooling down for 2 h at room temperature, the annealed probes were aliquoted for storage at −20 °C and thawed on ice before use. The isotope-labeled probe was end-labeled by using (α-32P)-dATP (PerkinElmer) and the Klenow fragment of DNA polymerase I (Promega). The labeled probe was purified with Sephadex G-50.
Binding reaction mixtures contained 20 fmol of the DNA biotin-labeled probe, various amounts of MBP-XerR or MBP-XccR protein in a buffer of 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 1 mM DTT, 50 ng/μl poly(dI-dC) DNA in a volume of 20 μl. After a 30-min incubation at room temperature, 4 μl of 80% glycerol was added to each reaction and samples were size fractionated using 5% polyacrylamide gels in 0.5× TBE buffer (45 mM Tris-borate; 1 mM EDTA) at 4 °C. For competition, a certain amount of unlabeled probe or plant signal(s) was co-incubated with the protein for 20 min at room temperature before adding labeled probe. The reaction samples were electrophoretically transferred to a nylon membrane (Hybond-N+, Amersham Biosciences) using wet transfer, and then the membranes were crosslinked by a UV lamp at 120 mJ/cm2. Detection of biotin-activated light signals was performed according to the manufacturer's instructions described by the LightShift Chemiluminescent EMSA Kit (Pierce).
For EMSA using isotope-labeled probe, 8 fmol of labeled probe was added to the mixtures. The reaction procedure and electrophoresis were the same as described above. The gel was dried and subjected to autoradiography.
Phosphorylation of MBP-XerR protein
Phosphorylation of the purified MBP-XerR protein was performed essentially as described previously 58, 59. Briefly, ∼50 μg of MBP-XerR was incubated with 50 mM acetyl phosphate (lithium, potassium salt, from Sigma) for 1 h at 30 °C in a buffer of 100 mM Tris-HC1 (pH 7.4), 10 mM MgC12, 125 mM KC1. The concentration of the phosphorylated protein was measured, and the conditions for using it in EMSA were the same as described above. In parallel, similar reactions lacking acetyl phosphate were used to prepare MBP-XerR for EMSA studies.
Preparation of low-molecular-weight plant extracts
About 20 g fresh cabbage leaves were homogenized in liquid nitrogen, and the powder was resuspended in 100 ml water. The extract was centrifuged, fractionated in series by 0.45 μm filter membrane, ultrafiltration membranes YM10 and YM1 to obtain the compounds of molecular weights less than 1 kDa.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No 31030008 and No 30471135), and the National Basic Research Program of China (2011CB100700).
References
- Decho AW, Norman RS, Visscher PT. Quorum sensing in natural environments: emerging views from microbial mats. Trends Microbiol. 2010;18:73–80. doi: 10.1016/j.tim.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Mullard A. Microbiology: Tinker, bacteria, eukaryote, spy. Nature. 2009;459:159–161. doi: 10.1038/459159a. [DOI] [PubMed] [Google Scholar]
- Antunes LC, Ferreira RB. Intercellular communication in bacteria. Crit Rev Microbiol. 2009;35:69–80. doi: 10.1080/10408410902733946. [DOI] [PubMed] [Google Scholar]
- Engebrecht J, Nealson K, Silverman M. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell. 1983;32:773–781. doi: 10.1016/0092-8674(83)90063-6. [DOI] [PubMed] [Google Scholar]
- Miller MB, Bassler BL. Quorum sensing in bacteria. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- Bassler BL, Losick R. Bacterially speaking. Cell. 2006;125:237–246. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Yang M, Zheng H, et al. Complex quorum-sensing regulatory systems regulate bacterial growth and symbiotic nodulation in Mesorhizobium tianshanense. Arch Microbiol. 2009;191:283–289. doi: 10.1007/s00203-008-0454-7. [DOI] [PubMed] [Google Scholar]
- Chevrot R, Rosen R, Haudecoeur E, et al. GABA controls the level of quorum-sensing signal in Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2006;103:7460–7464. doi: 10.1073/pnas.0600313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudecoeur E, Planamente S, Cirou A, et al. Proline antagonizes GABA-induced quenching of quorum-sensing in Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2009;106:14587–14592. doi: 10.1073/pnas.0808005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case RJ, Labbate M, Kjelleberg S. AHL-driven quorum-sensing circuits: their frequency and function among the Proteobacteria. ISME J. 2008;2:345–349. doi: 10.1038/ismej.2008.13. [DOI] [PubMed] [Google Scholar]
- Subramoni S, Venturi V. LuxR-family 'solos': bachelor sensors/regulators of signalling molecules. Microbiology. 2009;155:1377–1385. doi: 10.1099/mic.0.026849-0. [DOI] [PubMed] [Google Scholar]
- Patankar AV, Gonzalez JE. Orphan LuxR regulators of quorum sensing. FEMS Microbiol Rev. 2009;33:739–756. doi: 10.1111/j.1574-6976.2009.00163.x. [DOI] [PubMed] [Google Scholar]
- Lequette Y, Lee JH, Ledgham F, Lazdunski A, Greenberg EP. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J Bacteriol. 2006;188:3365–3370. doi: 10.1128/JB.188.9.3365-3370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danino VE, Wilkinson A, Edwards A, Downie JA. Recipient-induced transfer of the symbiotic plasmid pRL1JI in Rhizobium leguminosarum bv. viciae is regulated by a quorum-sensing relay. Mol Microbiol. 2003;50:511–525. doi: 10.1046/j.1365-2958.2003.03699.x. [DOI] [PubMed] [Google Scholar]
- McIntosh M, Krol E, Becker A. Competitive and cooperative effects in quorum-sensing-regulated galactoglucan biosynthesis in Sinorhizobium meliloti. J Bacteriol. 2008;190:5308–5317. doi: 10.1128/JB.00063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmer BM. Cell-to-cell signalling in Escherichia coli and Salmonella enterica. Mol Microbiol. 2004;52:933–945. doi: 10.1111/j.1365-2958.2004.04054.x. [DOI] [PubMed] [Google Scholar]
- Ryan RP, Dow JM. Diffusible signals and interspecies communication in bacteria. Microbiology. 2008;154:1845–1858. doi: 10.1099/mic.0.2008/017871-0. [DOI] [PubMed] [Google Scholar]
- Zhang L, Jia Y, Wang L, Fang R. A proline iminopeptidase gene upregulated in planta by a LuxR homolog is essential for pathogenicity of Xanthomonas campestris pv. campestris. Mol Microbiol. 2007;65:121–136. doi: 10.1111/j.1365-2958.2007.05775.x. [DOI] [PubMed] [Google Scholar]
- Ferluga S, Venturi V. OryR is a LuxR-family protein involved in interkingdom signaling between pathogenic Xanthomonas oryzae pv. oryzae and rice. J Bacteriol. 2009;191:890–897. doi: 10.1128/JB.01507-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferluga S, Bigirimana J, Hofte M, Venturi V. A LuxR homologue of Xanthomonas oryzae pv. oryzae is required for optimal rice virulence. Mol Plant Pathol. 2007;8:529–538. doi: 10.1111/j.1364-3703.2007.00415.x. [DOI] [PubMed] [Google Scholar]
- De Carlo S, Chen B, Hoover TR, et al. The structural basis for regulated assembly and function of the transcriptional activator NtrC. Genes Dev. 2006;20:1485–1495. doi: 10.1101/gad.1418306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Lee MA, Chun SJ, Park SJ, Lee KH. Role of NtrC in biofilm formation via controlling expression of the gene encoding an ADP-glycero-manno-heptose-6-epimerase in the pathogenic bacterium, Vibrio vulnificus. Mol Microbiol. 2007;63:559–574. doi: 10.1111/j.1365-2958.2006.05527.x. [DOI] [PubMed] [Google Scholar]
- Freeman JA, Bassler BL. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol Microbiol. 1999;31:665–677. doi: 10.1046/j.1365-2958.1999.01208.x. [DOI] [PubMed] [Google Scholar]
- Andrade MO, Alegria MC, Guzzo CR, et al. The HD-GYP domain of RpfG mediates a direct linkage between the Rpf quorum-sensing pathway and a subset of diguanylate cyclase proteins in the phytopathogen Xanthomonas axonopodis pv citri. Mol Microbiol. 2006;62:537–551. doi: 10.1111/j.1365-2958.2006.05386.x. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- Zhu X, Amsler CD, Volz K, Matsumura P. Tyrosine 106 of CheY plays an important role in chemotaxis signal transduction in Escherichia coli. J Bacteriol. 1996;178:4208–4215. doi: 10.1128/jb.178.14.4208-4215.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flashner Y, Weiss DS, Keener J, Kustu S. Constitutive forms of the enhancer-binding protein NtrC: evidence that essential oligomerization determinants lie in the central activation domain. J Mol Biol. 1995;249:700–713. doi: 10.1006/jmbi.1995.0330. [DOI] [PubMed] [Google Scholar]
- Skerker JM, Perchuk BS, Siryaporn A, et al. Rewiring the specificity of two-component signal transduction systems. Cell. 2008;133:1043–1054. doi: 10.1016/j.cell.2008.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, De La Torre A, Yan D, et al. Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains. Genes Dev. 2003;17:2552–2563. doi: 10.1101/gad.1125603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo F. Mechanisms of transcriptional repression. Curr Opin Microbiol. 2001;4:145–151. doi: 10.1016/s1369-5274(00)00180-6. [DOI] [PubMed] [Google Scholar]
- Rojo F. Repression of transcription initiation in bacteria. J Bacteriol. 1999;181:2987–2991. doi: 10.1128/jb.181.10.2987-2991.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker CT, Sperandio V. Cell-to-cell signalling during pathogenesis. Cell Microbiol. 2009;11:363–369. doi: 10.1111/j.1462-5822.2008.01272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Bodman SB, Bauer WD, Coplin DL. Quorum sensing in plant-pathogenic bacteria. Annu Rev Phytopathol. 2003;41:455–482. doi: 10.1146/annurev.phyto.41.052002.095652. [DOI] [PubMed] [Google Scholar]
- Studholme DJ, Dixon R. Domain architectures of σ54-dependent transcriptional activators. J Bacteriol. 2003;185:1757–1767. doi: 10.1128/JB.185.6.1757-1767.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern D, Volkman BF, Luginbuhl P, et al. Structure of a transiently phosphorylated switch in bacterial signal transduction. Nature. 1999;402:894–898. doi: 10.1038/47273. [DOI] [PubMed] [Google Scholar]
- Wyman C, Rombel I, North AK, Bustamante C, Kustu S. Unusual oligomerization required for activity of NtrC, a bacterial enhancer-binding protein. Science. 1997;275:1658–1661. doi: 10.1126/science.275.5306.1658. [DOI] [PubMed] [Google Scholar]
- Moore JB, Shiau SP, Reitzer LJ. Alterations of highly conserved residues in the regulatory domain of nitrogen regulator I (NtrC) of Escherichia coli. J Bacteriol. 1993;175:2692–2701. doi: 10.1128/jb.175.9.2692-2701.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombel I, North A, Hwang I, Wyman C, Kustu S. The bacterial enhancer-binding protein NtrC as a molecular machine. Cold Spring Harb Symp Quant Biol. 1998;63:157–166. doi: 10.1101/sqb.1998.63.157. [DOI] [PubMed] [Google Scholar]
- Boehr DD. During transitions proteins make fleeting bonds. Cell. 2009;139:1049–1051. doi: 10.1016/j.cell.2009.11.031. [DOI] [PubMed] [Google Scholar]
- Gardino AK, Villali J, Kivenson A, et al. Transient non-native hydrogen bonds promote activation of a signaling protein. Cell. 2009;139:1109–1118. doi: 10.1016/j.cell.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney LJ. How important is the phosphatase activity of sensor kinases. Curr Opin Microbiol. 2010;13:168–176. doi: 10.1016/j.mib.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silversmith RE. Auxiliary phosphatases in two-component signal transduction. Curr Opin Microbiol. 2010;13:177–183. doi: 10.1016/j.mib.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter SL, Roberts MA, Manning CS, Armitage JP. A bifunctional kinase-phosphatase in bacterial chemotaxis. Proc Natl Acad Sci USA. 2008;105:18531–18536. doi: 10.1073/pnas.0808010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton JA, Fray RG. Integration of environmental and host-derived signals with quorum sensing during plant-microbe interactions. Cell Microbiol. 2004;6:213–224. doi: 10.1111/j.1462-5822.2004.00362.x. [DOI] [PubMed] [Google Scholar]
- Piper KR, Beck Von Bodman S, Hwang I, Farrand SK. Hierarchical gene regulatory systems arising from fortuitous gene associations: controlling quorum sensing by the opine regulon in Agrobacterium. Mol Microbiol. 1999;32:1077–1089. doi: 10.1046/j.1365-2958.1999.01422.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Jiang G, Cui Y, et al. kdgREcc negatively regulates genes for pectinases, cellulase, protease, harpinEcc, and a global RNA regulator in Erwinia carotovora subsp. carotovora. J Bacteriol. 1999;181:2411–2421. doi: 10.1128/jb.181.8.2411-2421.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight PD, Kolter R. Interspecies chemical communication in bacterial development. Annu Rev Microbiol. 2009;63:99–118. doi: 10.1146/annurev.micro.091208.073248. [DOI] [PubMed] [Google Scholar]
- Manefield M, Rasmussen TB, Henzter M, et al. Halogenated furanones inhibit quorum sensing through accelerated LuxR turnover. Microbiology. 2002;148:1119–1127. doi: 10.1099/00221287-148-4-1119. [DOI] [PubMed] [Google Scholar]
- Rajamani S, Bauer WD, Robinson JB, et al. The vitamin riboflavin and its derivative lumichrome activate the LasR bacterial quorum-sensing receptor. Mol Plant Microbe Interact. 2008;21:1184–1192. doi: 10.1094/MPMI-21-9-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto MJ, Dominguez-Ferreras A, Perez-Mendoza D, Sanjuan J, Olivares J. Mutualism versus pathogenesis: the give-and-take in plant-bacteria interactions. Cell Microbiol. 2009;11:381–388. doi: 10.1111/j.1462-5822.2009.01282.x. [DOI] [PubMed] [Google Scholar]
- Yang Z, Rogers LM, Song Y, Guo W, Kolattukudy PE. Homoserine and asparagine are host signals that trigger in planta expression of a pathogenesis gene in Nectria haematococca. Proc Natl Acad Sci USA. 2005;102:4197–4202. doi: 10.1073/pnas.0500312102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner P, Barber C, Daniels M. Behavior of the transposons Tn5 and Tn7 in Xanthomonas campestris pv. campestris. Mol Gen Genet. 1984;195:101–107. [Google Scholar]
- Schafer A, Tauch A, Jager W, et al. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145:69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innes RW, Hirose MA, Kuempel PL. Induction of nitrogen-fixing nodules on clover requires only 32 kilobase pairs of DNA from the Rhizobium trifolii symbiosis plasmid. J Bacteriol. 1988;170:3793–3802. doi: 10.1128/jb.170.9.3793-3802.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogliano J, Lynch AS, Belin D, Lin EC, Beckwith J. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 1997;11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- Lynch AS, Lin EC. Transcriptional control mediated by the ArcA two-component response regulator protein of Escherichia coli: characterization of DNA binding at target promoters. J Bacteriol. 1996;178:6238–6249. doi: 10.1128/jb.178.21.6238-6249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]