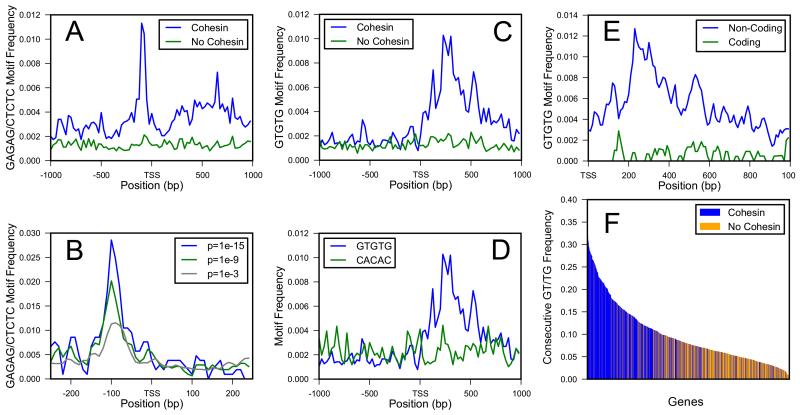
Figure 1.
Enrichment of GAF Binding Sites and GT Repeats in Cohesin-Binding Genes
(A) Combined frequency of GAGAG and CTCTC, binding sequences for GAGA factor (GAF), at positions from −1 kb to +1 kb around the TSS of 506 cohesin-binding genes (blue) and 1040 genes that do not bind cohesin (green).
(B) GAGAG and CTCTC frequency increases with the stringency with which cohesin binding is called (blue, p ≤ 10−15; green, p ≤ 10−9; gray, p ≤ 10−3).
(C) Frequency of GTGTG from −1 kb to +1 kb in cohesin-binding (blue) and non-binding (green) genes.
(D) Frequency of GTGTG (blue) and CACAC (green) in cohesin-binding genes.
(E) Frequency of GTGTG in non-coding (blue) and coding (green) sequences of cohesin-binding genes.
(F) Frequency of a GT or TG dinucleotide sequence following another in the 506 cohesin-binding (blue) and 1040 non-binding genes (yellow-orange) in the 1 kb region downstream of the TSS. The genes are in order of decreasing consecutive dinucleotide frequency.
Comparison of genome-wide binding of Nipped-B to GAGA factor (GAF) in Figure S1 indicates that the GAGAG sequences enriched in cohesin-binding genes are functional binding sites.