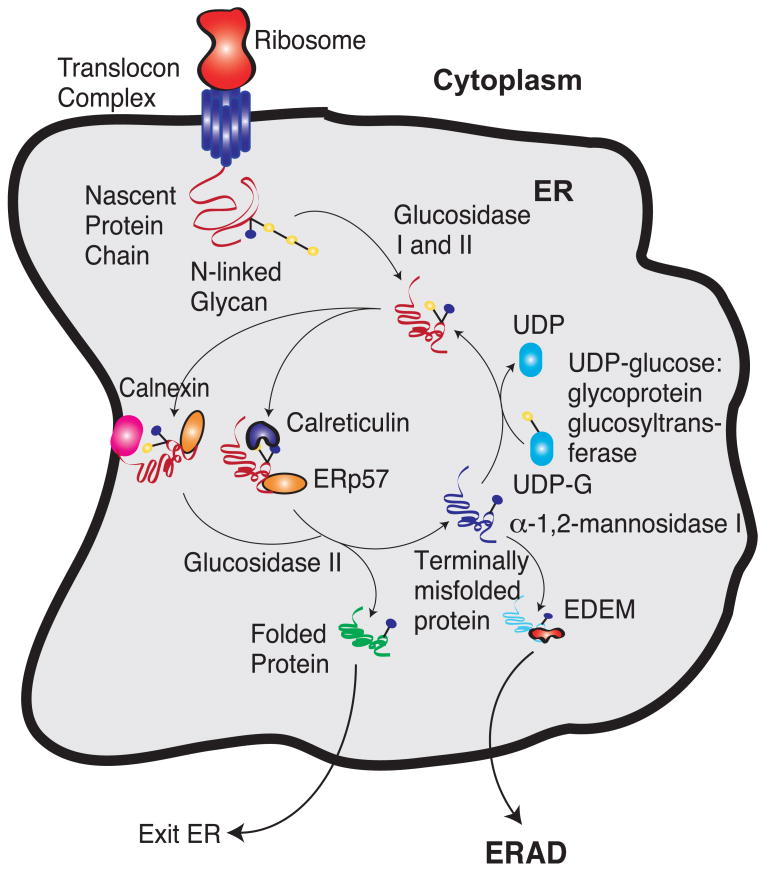
Fig. 1.
The calnexin/calreticulin protein folding cycle. Yellow circles denote glucose groups and while blue circles denote mannose groups. After entering the ER lumen, glucosidase I and II remove two glucose groups. The monoglucosylated glycoprotein then interacts with calnexin/calreticulin. These chaperones interact with the thiol-disulphide oxidoreductase ERp57. Cleavage of the last glucose residue by glucosidase II leads to the release of the chaperones. At this time, the protein could have either folded and left the ER or it could have attained an incorrect state. The incorrectly folded proteins are then the substrates of UDP glucose:glycoprotein glucosyltransferase, which puts a glucose residue back to the incorrectly folded protein. This enables the protein to spend some more time in folding in the ER. If the protein fails to fold in a repeated number of cycles, the mannose residue is removed by α-1,2-mannosidase I. This enables the protein to be recognized by ER-degradation-enhancing 1,2-mannosidase-like protein (EDEM). This targets the unfolded proteins for ER-associated degradation (ERAD).