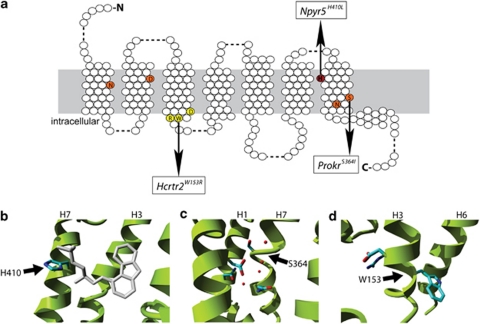
Figure 2.
Illustrations of the mutant structural environments in homology models of the mutated receptors. (a) Schematic overview of a consensus GPCR with the mutation shown in (b) in red, in orange the ionic pocket26 and mutated residue in (c) and in yellow the (D/E)R(Y/W) motif and mutated residue depicted in (d). (b) An example of a mutation that is predicted to affect ligand binding. The mutant H410L in the neuropeptide receptor NPY5R is located in the putative ligand-binding pocket. The structure of the co-crystallized ligand of the β2-adrenergic receptor is shown in gray. Although the NPY5R receptor binds a different class of ligands the binding site location is expected to be similar. Substituting the histidine for leucine is likely to change ligand-binding affinity. (c) The mutant S364I in the prokineticin 2 (PROK2) is located just above the ionic pocket, which is involved in signal transduction from the ligand-binding site to the G-protein-binding site. A number of structural waters are located in this pocket. The substitution of the serine for isoleucine is likely to disrupt the ionic pocket due to steric constraints, a major change in hydrophobicity and loss of interactions with structural waters. (d) The mutant W153R in the hypocretin (orexin) receptor 2 (HCRTR2) is located in the (D/E)R(Y/W) motif, which is the most conserved part of the GPCR family and involved in receptor activation and subsequent G-protein coupling. The substitution of trypthophan for arginine will disrupt receptor activation.