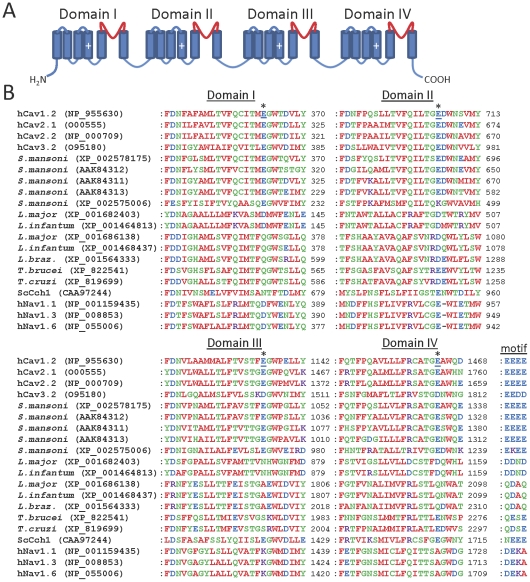
Figure 6. Parasite Cav channel homologues show similarity to human Cav channels in the pore region.
(A) Schematic showing the four-domain structure of human Cav channels, with the pore loop of each domain shown in red. Cylinders indicate TMDs, and plus signs indicate the charged voltage sensor regions. (B) Multiple sequence alignments of the pore domains of human Cav channels with parasite Cav channel homologues. Only those parasite homologues containing four putative domains are shown. Sequences of human Cav1.2 (L-type), Cav2.1 (P/Q-type), Cav2.2 (N-type) and Cav3.2 (T-type) channels, as well as the S. cerevisiae Cch1 Ca2+ channel (ScCch1), are shown. L. braz denotes L. braziliensis. Selected human Nav isoforms are included, to allow comparison with the related Cav channels. The locations of acidic residues (underlined) forming an acidic ring motif in human Cav1.2 channels are indicated by asterisks. The overall motif formed by all four domains at this locus is indicated to the right of the Domain IV alignment for each channel homologue.