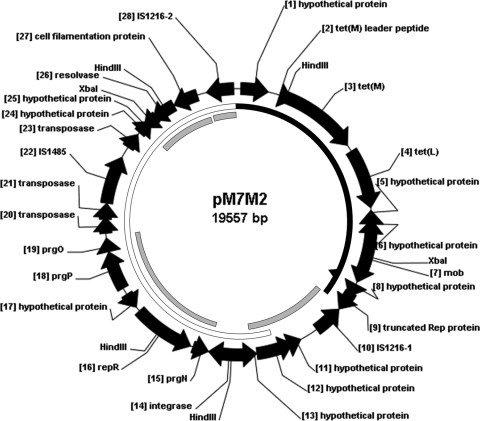
Fig. 1.
Circular map of plasmid pM7M2 (see Table 1 for further details). The putative open reading frames are numbered. The coding regions are represented by arrows indicating the direction of transcription, and putative proteins encoded by them correspond to the proteins from the database with the highest amino acid homology. Black solid ring, 99.5% nucleotide sequence identity (positions 1 to 7097) with S. gallolyticus UCN34 (GenBank accession no. FN597254.1) genome, which has an additional 32 bp behind position 6493 of pM7M2, as indicated by the black solid triangle; gray solid ring, 99% to 100% nucleotide sequence identity (positions 7098 to 9438, 10500 to 14353, 17014 to 18706, and 18749 to 19557) with E. faecium plasmid pIP816 (GenBank accession no. AM932524.1); open ring, 99.9% nucleotide sequence identity (positions 8744 to 19557) with E. faecalis plasmid pRE25 (GenBank accession no. X92945.2) and E. faecium plasmid p5753cB (GenBank accession no. GQ900487.1).