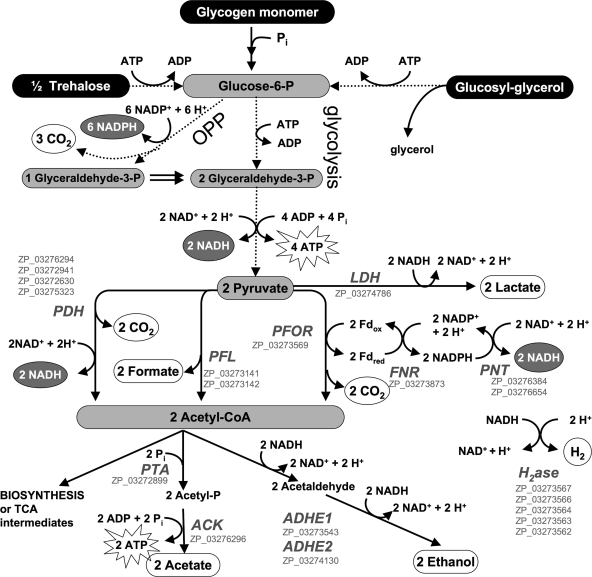
Fig. 1.
Catabolism of glucose equivalents derived from previously observed substrates (black ovals) (13), the experimentally observed excreted products (white ovals), and enzymes encoded in the A. maxima genome (gray italic characters) as well as their NCBI reference sequences (gray roman characters). All proteins necessary for these reactions are encoded in the A. maxima draft genome. Multistep reactions are abbreviated (indicated by dotted arrows without enzyme annotation). The OPP pathway abbreviated here was determined based on the idea of full cycling of fructose-6-phosphate. A more detailed scheme to illustrate the stoichiometry of OPP is provided in Fig. S1 in the supplemental material. Enzyme abbreviations: LDH, lactate dehydrogenase; PDH, pyruvate dehydrogenase; PFL, pyruvate formate lyase; PFOR, pyruvate ferredoxin oxidoreductase; FNR, ferredoxin-NADP reductase; PNT, pyridine-NADH transhydrogenase; PTA, phosphotransacetylase; ACK, acetate kinase; ADHE, bifunctional aldehyde alcohol dehydrogenase; H2ase, hydrogenase.