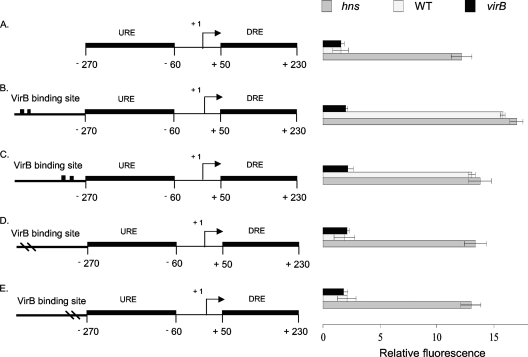
Fig. 2.
Derepression of the proU promoter by VirB. Summaries of the structures of various derivatives of the E. coli proU promoter region are presented at the left; levels of expression of the proU-gfp transcriptional reporter fusion are given at the right (diagrams not to scale). The native proU promoter is shown (A); proU incorporating functional VirB binding and nucleation sites (denoted by two black boxes) is shown in the forward (B) and reverse (C) orientations, and proU with inactivated VirB binding sites (denoted by two dashes in the VirB site) is shown in the forward (D) and reverse (E) orientations (drawings not to scale). The constructs were assessed in S. flexneri for proU-gfp expression in a virB mutant (black bars), the wild type (WT) (white bars), and the hns mutant (gray bars) under conditions normally repressive of proU transcription.