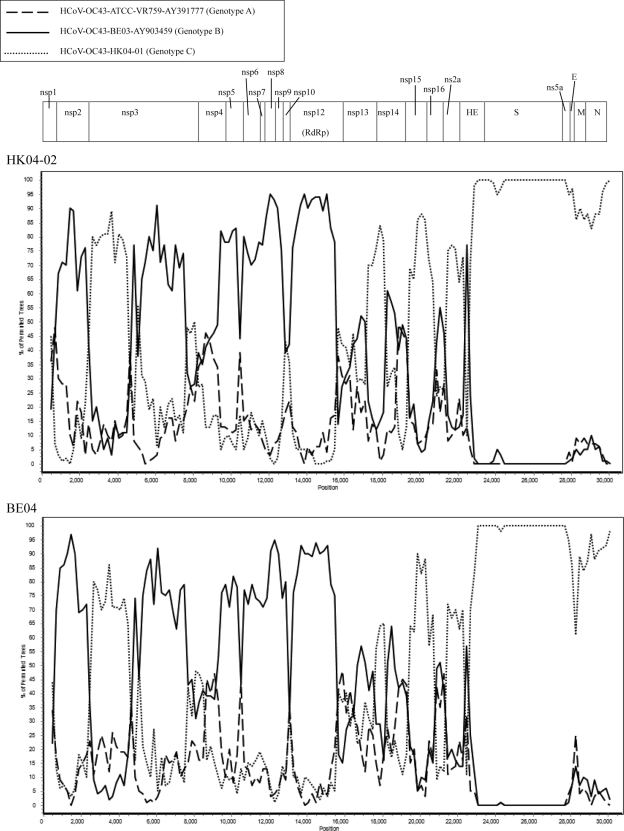
Fig. 2.
Genome organization and bootscan analysis of the HCoV-OC43 genomes. Bootscanning was conducted with Simplot version 3.5.1 (F84 model; window size, 1,000 bp; step, 200 bp) on a gapless nucleotide alignment, which was generated with ClustalX with the genome sequences of strains HK04-02 (upper) and BE04 (lower) as the query sequences.