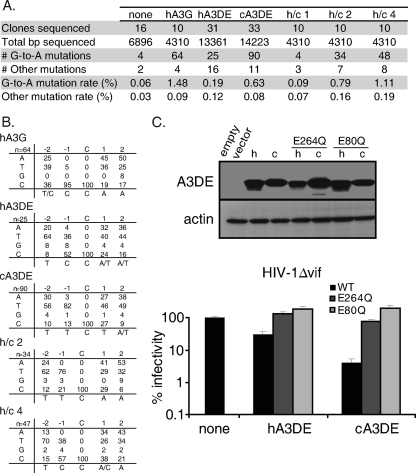
Fig. 6.
Chimpanzee Apobec3DE induces higher levels of hypermutation than human Apobec3DE does during viral infection. (A) HIV-1 genomes were sequenced from HIV-1Δvif infections performed in the absence of Apobec3 (none) or in the presence of human Apobec3G (hA3G), human Apobec3DE (hA3DE), chimpanzee Apobec3DE (cA3DE), or human/chimpanzee chimeras, and the G-to-A mutation rate of viral genomes was calculated. The rate of other mutations was also calculated. Combined data from three independent experiments are shown. (B) The nucleotide context of G-to-A mutations was characterized. The percentage of each nucleotide occurring at the −2, −1, +1, and +2 positions of deaminated cytidines is shown, with the most common nucleotide listed below each position. The number of mutations examined is noted in the top left corner of each grid. (C) (Top) Western blot analysis of cellular levels of human (h) and chimpanzee (c) Apobec3DE catalytic mutants. Apobec3DE was detected with an anti-HA antibody. Actin was used as a loading control. (Bottom) Infectivity of HIV-1Δvif in the presence of wild-type Apobec3DE (WT) or Apobec3DE catalytic mutants (E264Q and E80Q). Infections are represented as a percentage of the infectivity of HIV-1 without Apobec3 (none), which was set to 100%. Experiments were performed at least 2 times, and results from one representative experiment are shown. Error bars represent the standard deviation of triplicate infections within one experiment.