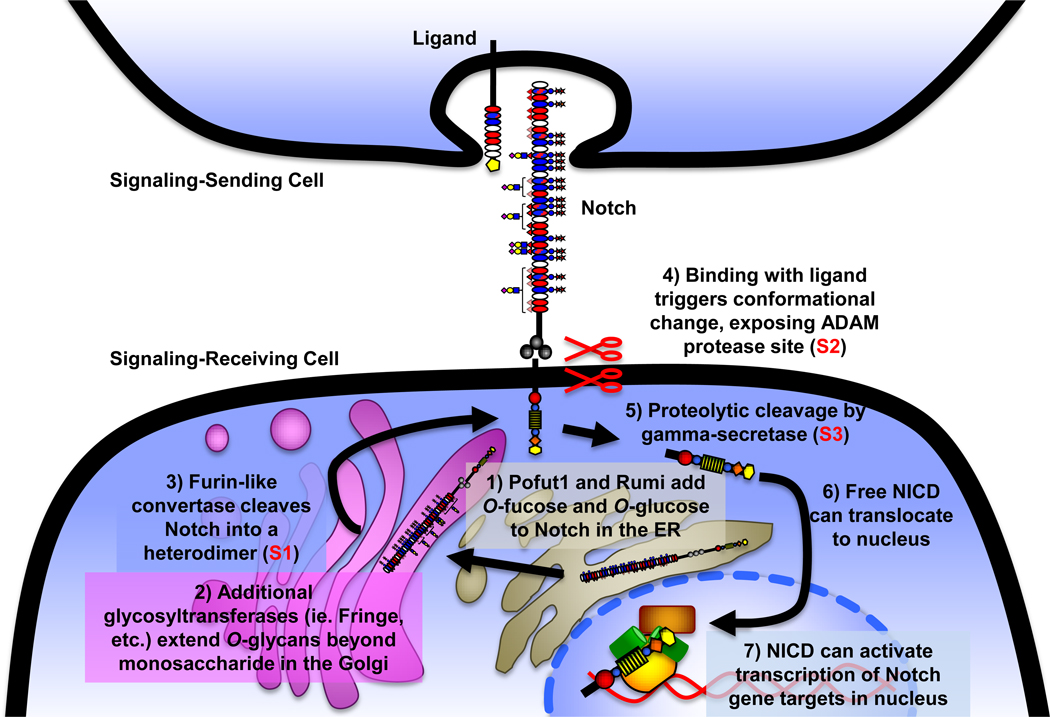
Figure 2. Notch activation pathway.
Notch is O-fucosylated and O-glucosylated in the ER (1), and elongated by additional glycosyltransferases in the Golgi (2). Notch is also cleaved by a furin-like convertase in the Golgi (S1 cleavage, 3), generating the mature heterodimeric receptor that is subsequently expressed at the cell surface. Following interaction with a DSL (Delta, Serrate, Lag2) ligand presented on an apposing cell, Notch ECD undergoes a conformational change exposing the protease site for an ADAM protease (Kuzbanian in Drosophila) at the membrane interface (S2 cleavage, 4), and is subsequently cleaved by gamma-secretase just inside the cell membrane (S3 cleavage, 5), causing ICD release. Upon S3 cleavage, the ICD is free to translocate to the nucleus (6), bind to a member of the CSL (CBF/SuH/LAG-1) family of transcription factors via its RAM domain, and this binary complex serves as a platform for Mastermind-like (MAML)-family cofactors, which in turn can act as scaffolding for transcriptional complex formation. The assembled transcriptional activation complex can then activate expression of Notch target genes (7), including Hairy-Enhancer of Split (Hes) and Hairy-Enhancer of Split related with YRPW motif (Hey) family members [1].