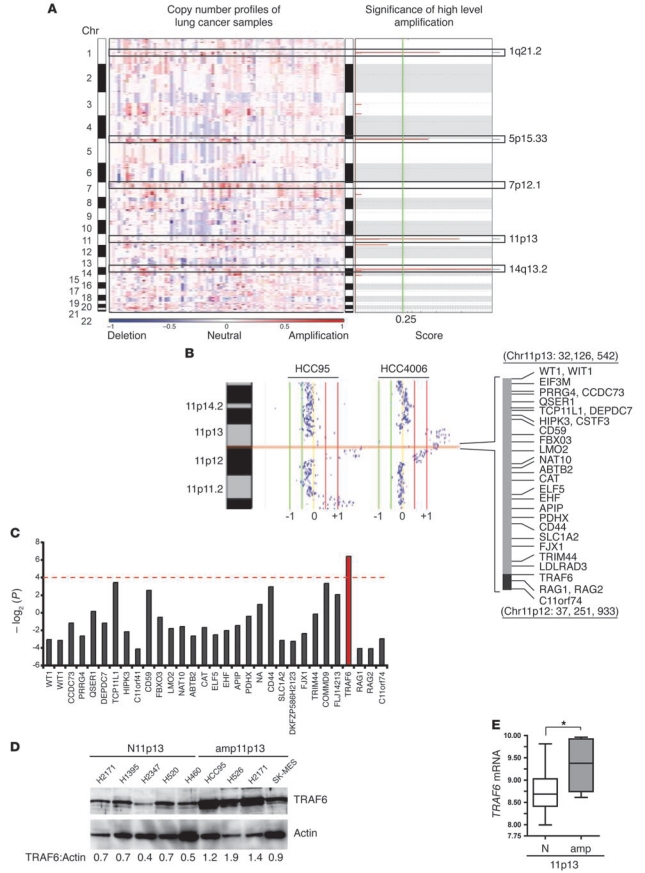
Figure 1. TRAF6 locus amplification occurs frequently in lung cancer.
(A) Copy number data for 85 lung cancer lines is shown by genomic location (rows). False discovery rates (q values; 0.25 cutoff for significance shown by green line) and scores for amplicons are plotted at each genome position. Individual chromosomes are denoted by white and gray shading; black boxes indicate significant high-level amplifications. (B) Copy number status of chromosome 11p for 2 representative lung cancer lines. The minimally amplified region spanning chromosome 11p13 to 11p12 (32,126,542–37,251,933) and known genes are shown at right. Normalized log2 signal intensity ratios were plotted and visualized by SIGMA2 (32). Vertical lines denote log2 signal ratios from –1 to +1; copy number gains (red) are to the right of 0. Each dot represents a single probe. (C) A subset of 66 lung cancer samples was analyzed to identify genes whose expression correlated with gene amplification. P values (–log2) for genes within the minimally amplified region on 11p13 are shown as a histogram. Values above the dashed line denote P ≤ 0.05. (D) IB for TRAF6 and actin in 5 lung cancer lines diploid at 11p13 (N, no 11p13 amplification; H2171, H1395, H2347, H520, and H460) and 4 lung cancer lines with an 11p13 amplification (amp; HCC95, H526, H2171, and SK-MES-1). Densitometric values for TRAF6 protein relative to actin are shown below. (E) TRAF6 mRNA levels were evaluated in lung tumor samples with (n = 13) and without (n = 31) amplification of 11p13. Significance was determined by Mann-Whitney U test. *P < 0.05.