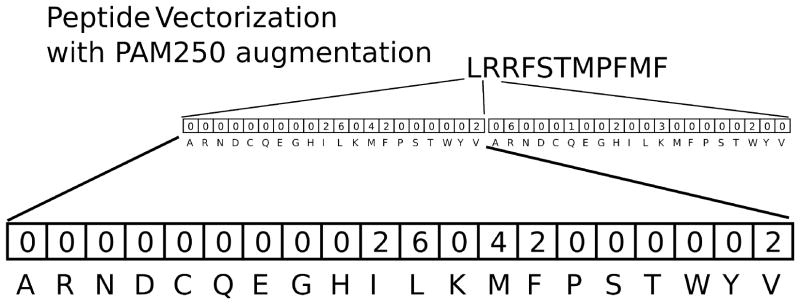
Figure 2. Peptide vectorization.
Each peptide is converted into a sparse vector which uniquely maps specific amino acids to positions in the peptide. The mapping is augmented by the PAM250 amino acid substation matrix. PAM matrices are based on the empirical mutation rate of amino acids in evolutionarily related proteins. For example, the figure shows the vectorization of the peptide LRRFSTMPFMF. The first amino acid leucine (L) can mutate to isoleucine (I), methionine (M), phenylalanine (F), and valine (V) at a rates greater than expected by chance. The weights assigned to these amino acids are given by log odds ratio in the PAM250 matrix. All other amino acids mutate from leucine at a lower rate than expected by chance. As a result, their value is set to zero. The PAM matrix gave us a principled way to associate common amino acids based on their chemical and structural properties.