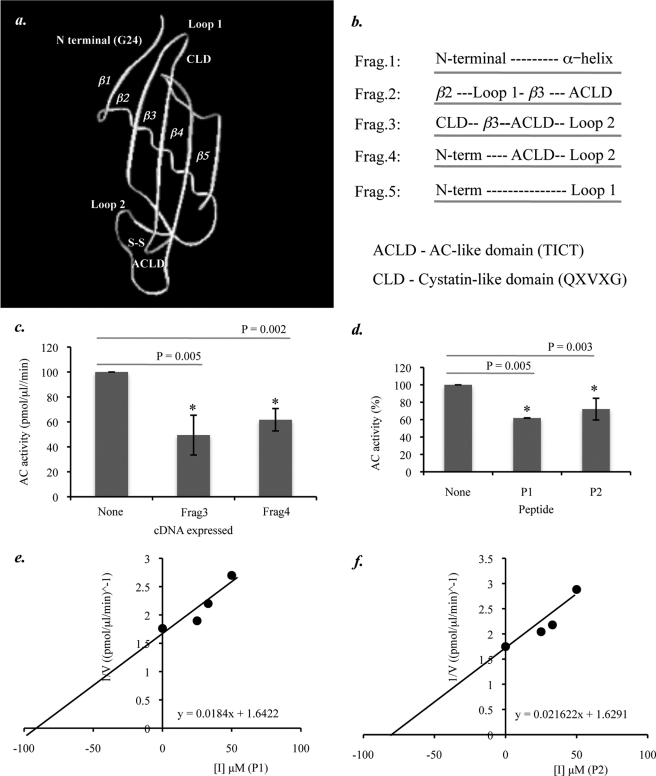
FIGURE 5.
Computer modeling and construction of cysSA-based peptides. a, shown is a proposed model of cysSA, generated by 3D-JIGSAW protein homology modeling based on PDB entry 1rn7, showing the location of potential functional domains (i.e. CLD; QXVXG), the N-terminal region, Loop 1, Loop 2, and ACLD (TICT). b, shown is the design of partial cDNA fragments encoding the potential functional domains of cysSA. c, AC activity was measured in HEK 293T17 cell extracts after transient expression of the AC cDNA and co-expression of Fragments 1–5, subcloned into the pCMV vector. AC activity in the presence of Fragments 2 and 3, both of which contain the AC-like domain, was significantly lower in comparison to AC only (t test, p = 0.005 and 0.002, respectively). Other peptides did not have a significant effect on AC activity (data not shown). Data represent the mean ± S.E., n = 3 independent experiments. d, the activity of pure, recombinant AC was determined in the presence of three different amounts (25, 33, and 50 μm) of peptides P1 and P2 as well as a nonspecific control peptide (Pc) using 0.1 μm NBD-C12 ceramide. The relative AC activity, averaged for all peptides concentrations, was significantly reduced for both of the cysSA-based peptides (p = 0.005 and 0.003, respectively) but not for the control peptide. e and f, Dixon plots (1/V versus [I]) were used to estimate the Ki values for P1 (e) and P2 (f) though equations (y = 0.0184x + 1.6422 and y = 0.021622x + 1.6291, respectively). Data represent the mean ± S.E., n = 3 independent experiments.