FIGURE 3.
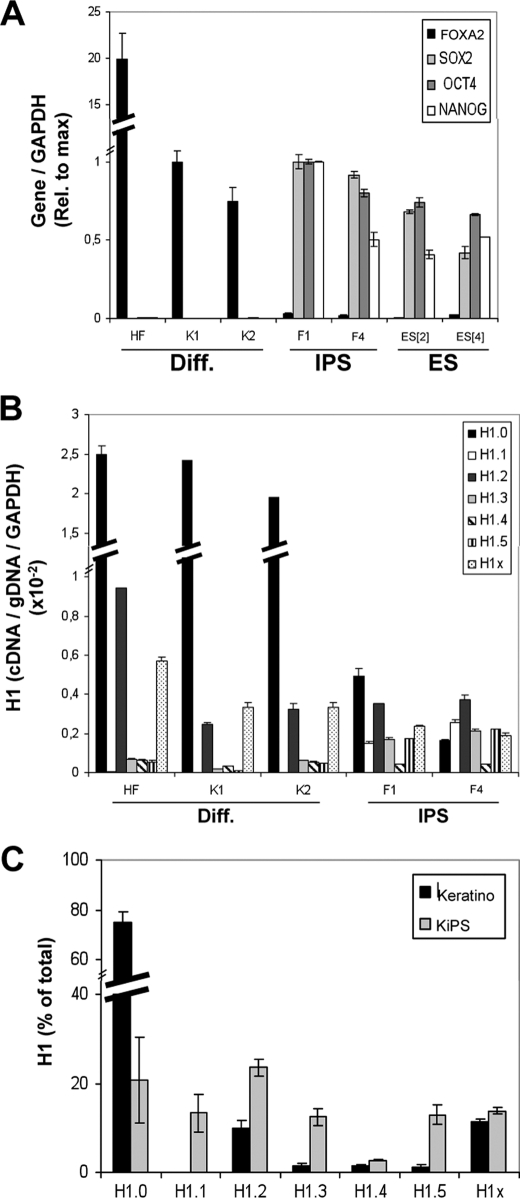
Changes of histone H1 variant gene expression upon reprogramming of human keratinocytes to iPS cells. A, expression of pluripotency or differentiation markers (Diff.) in normal and reprogrammed keratinocytes. RNA was extracted from human cell lines derived from HF, keratinocytes (K1 and K2), iPS cells obtained by reprogramming of keratinocytes (KiPS4F1 and KiPS4F4; named F1 and F4 in the figures), and hES cells (ES[2] and ES[4]) and used to determine expression of FOXA2, SOX2, OCT4, and NANOG by RT-qPCR with specific primer pairs. GAPDH expression was measured for normalization. Expression of each gene relative to the maximal value in keratinocytes (FOXA2) or iPS cells (SOX2, OCT4, and NANOG) is represented. B, histone H1 variant gene expression in normal and reprogrammed keratinocytes. The RNA samples of HF, keratinocytes (K1 and K2) and iPS cells (KiPS4F1 and KiPS4F4) were used to measure histone H1 expression by RT-qPCR with variant-specific oligonucleotide pairs. Histone H1 variant cDNA amplification was normalized with the values of HF or K1-derived genomic DNA amplified with the same primer set and GAPDH expression, as described in Fig. 1B. C, proportions of histone H1 variant transcripts in normal and reprogrammed keratinocytes. The relative proportion of each H1 variant expression in keratinocytes and iPS cells was calculated from the data in B and expressed as a percentage of the total H1 content. The data are expressed as the mean values for K1 and K2 (keratinocytes) or KiPS4F1 and KiPS4F4 (KiPS), respectively. The means and S.D. of a representative experiment quantified in triplicate are shown.