Background: Two-component signal transduction systems regulate important physiological functions in bacteria.
Results: The Fe(III), Fe(II)-O2, and Fe(II)-CO complexes of a heme-bound globin-coupled histidine kinase from Anaeromyxobacter displayed autophosphorylation activity, whereas the Fe(II) complex was inactive.
Conclusion: Gas binding and heme redox regulate the histidine kinase function.
Significance: A novel heme-based globin-coupled oxygen sensor histidine kinase was identified and characterized.
Keywords: Bacterial Protein Kinases, Enzyme Catalysis, Heme, Histidine Kinases, Oxygen Binding, Raman Spectroscopy, Redox Signaling, Signal Transduction, Spectroscopy, Oxygen Sensor
Abstract
Two-component signal transduction systems regulate numerous important physiological functions in bacteria. In this study we have identified, cloned, overexpressed, and characterized a dimeric full-length heme-bound (heme:protein, 1:1 stoichiometry) globin-coupled histidine kinase (AfGcHK) from Anaeromyxobacter sp. strain Fw109-5 for the first time. The Fe(III), Fe(II)-O2, and Fe(II)-CO complexes of the protein displayed autophosphorylation activity, whereas the Fe(II) complex had no significant activity. A H99A mutant lost heme binding ability, suggesting that this residue is the heme proximal ligand. Moreover, His-183 was proposed as the autophosphorylation site based on the finding that the H183A mutant protein was not phosphorylated. The phosphate group of autophosphorylated AfGcHK was transferred to Asp-52 and Asp-169 of a response regulator, as confirmed from site-directed mutagenesis experiments. Based on the amino acid sequences and crystal structures of other globin-coupled oxygen sensor enzymes, Tyr-45 was assumed to be the O2 binding site at the heme distal side. The O2 dissociation rate constant, 0.10 s−1, was substantially increased up to 8.0 s−1 upon Y45L mutation. The resonance Raman frequencies representing νFe-O2 (559 cm−1) and νO-O (1149 cm−1) of the Fe(II)-O2 complex of Y45F mutant AfGcHK were distinct from those of the wild-type protein (νFe-O2, 557 cm−1; νO-O, 1141 cm−1), supporting the proposal that Tyr-45 is located at the distal side and forms hydrogen bonds with the oxygen molecule bound to the Fe(II) complex. Thus, we have successfully identified and characterized a novel heme-based globin-coupled oxygen sensor histidine kinase, AfGcHK, in this study.
Introduction
Two-component signal transduction systems are involved in a wide variety of responses to environmental cues in bacteria (1–3). Typical two-component systems comprise a sensor histidine kinase and response regulator (RR)4 protein. Generally, sensor kinases are multidomain proteins containing a non-conserved sensory input domain responsible for detecting a particular stimulus or ligand and a conserved kinase domain (1–4). Similarly, RRs are multidomain proteins with a conserved receiver domain and variable output domain (5–7). In histidine kinases, a conserved His residue is initially autophosphorylated. The phosphate group is subsequently transferred to a conserved Asp in the receiver domain of the cognate RR, resulting in activation of the output domain.
Hitherto, several heme-regulated oxygen sensor histidine kinases have been characterized (8–19). In the root nodule bacteria, Rhizobium meliloti and Bradyrhizobium japonicum, FixLs with a heme-containing PAS domain regulate the expression of genes involved in nitrogen fixation and the anaerobic respiratory chain (11, 13). DevS (DosS) and DosT, containing a heme-bound GAF domain, regulate entry of the pathogenic bacterium, Mycobacterium tuberculosis, into dormancy (14–17). The PAS and GAF domains share structural similarities. These kinases are also designated heme-bound oxygen sensor enzymes, as association/dissociation to/from the heme iron complex in the sensor domain regulates their functions. The bacterial oxygen and NO sensor, H-NOX, is possibly a heme-regulated histidine kinase (18). In Corynebacterium diphtheriae, ChrS is a heme sensor histidine kinase involved in acquisition of host heme iron (19).
In addition to heme-regulated oxygen sensor histidine kinases with the PAS, GAF, and H-NOX domains, other types of oxygen sensor proteins with different functions, such as heme-regulated oxygen sensor phosphodiesterases with the PAS domain, EcDOS and AxPDEA1, have been identified (10, 13, 20–22). Moreover, an oxygen sensor protein, DcrA, with covalently bound c-type heme (23) and those with noncovalently bound b-type heme in the vitamin B12 binding domain, such as SCHIC (24), or in a globin fold have been documented. The latter enzyme is designated globin-coupled oxygen sensor (GCS) protein (25, 26). GCS proteins contain a heme-bound oxygen sensor domain with the globin fold in the N terminus. The structures of the globin fold of GCS enzymes are similar to those of myoglobin and hemoglobin but lack the entire D- and half of the E-helices of these proteins. Six heme-based oxygen sensor proteins, YddV (27, 28), HemAT-Bs (29–32), AvGReg (33), BpeGReg (34), GsGCS (35), and HemDGC (36), with the globin fold have been identified to date. Although both the sensor histidine kinase in the two-component system and GCS protein are important for bacteria to survive and/or to accommodate various stresses, no globin-coupled heme-regulated histidine kinases (GcHK) have been documented to date.
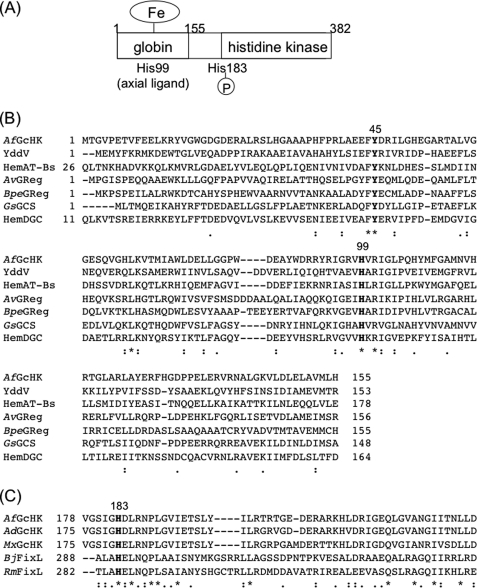
Based on sequence homology of the heme-bound globin domains of GCS proteins, including YddV, HemAT, and GRegs (25–36), we speculated on the presence of a novel sensor histidine kinase in soil slime bacteria, Anaeromyxobacter sp. strain Fw109-5, containing a globin sensor domain at the N terminus and histidine kinase domain at the C terminus. We designated this gene, Anae109_2438, as gchK (globin-coupled histidine kinase). The globin domain of the GcHK protein should bind heme, in view of its similarity to the amino acid sequences of the globin fold proteins, YddV, HemAT, GRegs, and HemDGC, which are heme-based globin-coupled oxygen sensor enzymes with sufficient heme binding affinity (25–36) (Fig. 1, A and B). The putative structure of the heme binding site was conjectured based on the structure of the cyanide-bound complex of HemAT-Bs (32). The heme binding residue located at the proximal side was identified as His-99, and Tyr-45 at the heme distal side appeared critical in oxygen binding (Fig. 1, A and B) (28, 31, 32, 36). Accordingly, it is reasonable to hypothesize that GcHK from the bacterial Anaeromyxobacter sp. strain Fw109-5 (AfGcHK) is a novel globin-coupled heme-based oxygen sensor protein displaying autophosphorylation activity in response to oxygen availability.
FIGURE 1.
A, the putative domain structure of AfGcHK is shown. The heme binding site is suggested as His-99 in the globin domain at the N terminus, and the autophosphorylation site is suggested as His-183 near the central region. B, amino acid alignment of the globin domain of AfGcHK and relevant globin-coupled oxygen sensor proteins are shown. It is assumed that Tyr-45 (Tyr-70, HemAT-Bs) is located at the heme distal side, whereas His-99 (His-123, HemAT-Bs) is the proximal axial ligand in AfGcHK. C, shown are amino acid sequences of the putative autophosphorylation sites of GcHK homologs and another histidine kinases, FixLs. His-183 is conserved among the histidine kinases. AdGcHK, a GcHK from A. dehalogenans; MxGcHK, a GcHK from M. xanthus.
In this study we identified, cloned, overexpressed, and purified AfGcHK and characterized the catalytic and physicochemical properties of wild-type and mutant proteins for the first time. Purified AfGcHK was clearly a dimeric heme-binding protein. Significant autophosphorylation activities were observed for the Fe(III), Fe(II)-O2, and Fe(II)-CO complexes but not the Fe(II) complex. Data obtained on the physicochemical properties of the mutant enzymes suggested that His-99 is the heme axial ligand at the proximal side, and Tyr-45 located at the distal side forms hydrogen bonds with oxygen bound to the heme iron complex, whereas His-183 is an autophosphorylation site of AfGcHK. Kinase activity toward RR (phosphate transfer reaction from phosphorylated AfGcHK to two sites on RR) by the Fe(III) and Fe(II)-O2 forms of AfGcHK was confirmed, and the phosphorylation sites on RR were identified using site-directed mutagenesis experiments.
EXPERIMENTAL PROCEDURES
Materials
Hemin chloride was obtained from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). Other chemicals acquired from Wako Pure Chemical Industries (Osaka, Japan) were of the highest guaranteed grade available and were used without further purification. Oligonucleotides were synthesized by Operon Biotechnologies (Tokyo, Japan). Phos-tag acrylamide was from the Phos-tag consortium.
Construction of Expression Plasmids
cDNAs encoding Anae109_2438 (corresponding to AfGcHK) and Anae109_2439 (corresponding to RR) from Anaeromyxobacter sp. strain Fw109-5 were synthesized by Mr. Gene (Regensburg, Germany) and optimized for Escherichia coli expression. Anae109_2438 cDNA was digested with NdeI and XhoI and subcloned into the pET21c vector (Novagen, Darmstadt, Germany), leading to the introduction of a His6 tag at the C terminus of the desired protein. Anae109_2439 cDNA was digested with BamHI and EcoRI and subcloned into pGEX-6P-2 vector (GE Healthcare), introducing a glutathione S-transferase (GST) tag at the N terminus of the desired protein.
Site-directed Mutagenesis
Mutagenesis was conducted using the QuikChange mutagenesis kit from Stratagene and the PrimeSTAR mutagenesis basal kit from Takara Bio (Otsu, Japan). The presence of the desired mutations was confirmed via DNA sequencing by Greiner Bio-One (Tokyo, Japan). The oligonucleotides employed are listed in supplemental Table S1.
Overexpression and Purification of Full-length AfGcHK
Full-length wild-type and mutant AfGcHKs were expressed in E. coli BL21(DE3) (Novagen) harboring the pET21c(+) expression vector. Briefly, E. coli BL21(DE3) was transformed with the required plasmid, plated on LB agar containing 100 μg/ml ampicillin, and incubated at 37 °C overnight. The following day, a single colony was inoculated in LB containing 100 μg/ml ampicillin and shaken overnight at 200 rpm and 37 °C. The culture medium was added to Terrific Broth (1:1000 dilution) containing the above antibiotic and shaken at 120 rpm and 37 °C for 4 h. Subsequently, the medium was cooled to 15 °C, and protein expression was induced by adding 0.1 mm isopropyl β-d-thiogalactopyranoside followed by further shaking for 20 h. E. coli cells were harvested by centrifugation for 10 min at 6750 × g and 4 °C, frozen in liquid nitrogen, and stored at −80 °C until purification.
E. coli cells frozen at −80 °C were suspended in buffer A (50 mm Tris-HCl, pH 8.0, 100 mm NaCl), 1 mm phenylmethanesulfonyl fluoride, 1 mm EDTA, and 0.2 mg/ml lysozyme. The solution was sonicated, centrifuged at 100,000 × g for 30 min and incubated for 5 min with hemin (50 μm) in DMSO solution. Supernatant fractions were applied to the nickel-nitrilotriacetic acid-agarose (Qiagen, Hilden, Germany) column pre-equilibrated with buffer A containing 10 mm imidazole. Subsequently, the column was washed with buffer A containing 20 mm imidazole, and AfGcHK fractions were eluted with a linear gradient of 20–200 mm imidazole in buffer A. The AfGcHK-containing solution was dialyzed against 20 mm Tris-HCl, pH 8.0 (buffer B), and concentrated with Amicon Ultra (Millipore, Billerica, MA). Proteins were immediately frozen in liquid nitrogen and stored at −80 °C until use. Concentrations were determined using the Quick Start Bradford Protein Assay kit (Bio-Rad) for protein and the pyridine hemochromogen method for heme (37). Purified proteins were >90% homogenous, as confirmed using SDS-PAGE (supplemental Fig. S1).
Overexpression and Purification of GST-tagged RR Protein
Full-length wild-type and mutant GST-RR proteins were expressed in E. coli BL21(DE3) harboring the pGEX-6P-2 expression vector. E. coli cells were suspended in buffer C (10 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.5, 140 mm NaCl, 2.7 mm KCl) with 1 mm EDTA, 1 mm phenylmethanesulfonyl fluoride, and 0.2 mg/ml lysozyme. Cells were crushed via pulsed sonication for 2 min (3 times with 2-min intervals) on ice and centrifuged at 100,000 × g for 30 min at 4 °C. The supernatant factions were applied to a glutathione-Sepharose 4B column (GE Healthcare). Subsequently, the column was washed with buffer C, and protein fractions were eluted with 10 mm reduced glutathione in 50 mm Tris-HCl, pH 8.0. Fractions were pooled and dialyzed against 20 mm Tris-HCl, pH 8.0. Purified proteins were concentrated with Amicon Ultra device (Millipore), immediately frozen in liquid nitrogen, and stored at −80 °C until use. Purified GST-RR was more than 95% homogeneous, as determined with SDS-PAGE, followed by staining with Coomassie Brilliant Blue (supplemental Fig. S1).
Gel Filtration Chromatography
To determine the oligomerization state, gel filtration was carried out using the ÄKTA liquid chromatography system equipped with a Superdex 200 HR 10/30 column (GE Healthcare). The buffer used for gel filtration was 20 mm Tris-HCl, pH 8.0, 150 mm NaCl. Molecular weight was estimated from the correlation between molecular weight and elution volume of standard proteins using a gel filtration calibration kit (GE Healthcare).
Optical Absorption Spectra
Absorption spectral data were obtained under aerobic conditions using a UV-2550PC (Shimadzu, Kyoto, Japan) spectrophotometer. Anaerobic spectral experiments were conducted on a Shimadzu UV-1650PC spectrophotometer in a glove box with 90% N2 and 10% H2 gas (O2 concentrations less than 50 ppm) (28). After reduction of heme with sodium dithionite, excess dithionite was removed using a Sephadex G-25 column (GE Healthcare) in the glove box. To ensure that the appropriate temperature of the solution was maintained, the reaction mixture was incubated for 5 min before spectroscopic measurements. The Fe(II) complex was prepared in N2-saturated buffer (50 mm Tris-HCl, pH 8.0). Fe(II)-O2 and -CO complexes were prepared in O2- and CO-saturated buffers (50 mm Tris-HCl, pH 8.0), respectively. Gas-saturated solutions were obtained by bubbling buffers with the appropriate gas for at least 30 min at room temperature.
Enzymatic Assays
Autophosphorylation activity was assayed at 25 °C in a reaction mixture containing 50 mm Tris-HCl, pH 8.0, 50 mm KCl, 5 mm MgCl2, and 10 μm AfGcHK. The phosphotransfer reaction was assayed at 25 °C in the same buffer containing 10 μm AfGcHK and 10 μm GST-RR. The reaction mixture was preincubated for 5 min, and the reaction was initiated by adding 1 mm ATP at 25 °C.
At the indicated times the reaction was terminated by adding 2× termination buffer (125 mm Tris-HCl, pH 6.8, 4% SDS, 4% 2-mercaptoethanol, 20% glycerol, 0.004% bromphenol blue). Samples were loaded on a 7.5% SDS poly-acrylamide gel containing 50 μm Phos-tag acrylamide and 0.1 mm MnCl2. Phosphorylated protein interacted with the Phos-tag manganese complex, so that mobility was slower than that of phosphate-free protein (38, 39). Proteins were visualized with Coomassie Brilliant Blue R350 (GE Healthcare) staining. Gel images were acquired using LAS-3000 (Fujifilm, Tokyo, Japan) and quantified using Multi-Gauge software (Fujifilm).
Pulse Radiolysis
O2 and CO association rate constants were obtained using pulse radiolysis. Experiments were performed essentially as described previously (28, 40–42).
O2 and CO Dissociation Rate Constants
To determine the O2 dissociation rate constant, 2 μm O2-bound AfGcHK was mixed with anaerobic buffer containing 50 mm Tris-HCl, pH 8.0, and 5 mm dithionite in an RSP-1000 stopped-flow spectrometer (Unisoku, Osaka, Japan) at 25 °C. The CO dissociation rate constant was estimated by mixing 5 μm CO-bound AfGcHK with 0.1 mm potassium ferricyanide in 50 mm Tris-HCl, pH 8.0, using a MultiSpec-1500 (Shimadzu) photodiode array spectrometer equipped with a Peltier cell temperature controller at 25 °C (28, 43, 44).
Resonance Raman Spectra
Resonance Raman spectra were obtained with a single polychromator (SPEX500M; HORIBA Jobin Yvon, Longjumeau, France) equipped with a liquid nitrogen-cooled CCD detector (Spec-10:400BLN; Roper Scientific, Sarasota, FL) (12, 23, 30, 36). The excitation wavelength employed was 413.1 nm from a krypton ion laser (BeamLok 2060; Newport Corp. & Spectra Physics Lasers, Mountain View, CA). Laser power at the sample point was adjusted to 5 milliwatts for air-oxidized, dithionite-reduced, and O2-bound forms and 0.1 milliwatt for the CO-bound form to prevent photodissociation of CO. Raman shifts were calibrated with indene, CCl4, acetone, and an aqueous solution of ferrocyanide. The accuracy of the peak position of well defined Raman bands was ±1 cm−1. The protein concentration for resonance Raman experiments was ∼20 μm in 50 mm Tris-HCl, pH 8.0.
RESULTS
Identification of a Novel Histidine Kinase in the Anaeromyxobacter sp. Fw109-5 Genome
Gene analysis of Anae109_2438 revealed the presence of a GCS with a globin domain at the N terminus and a histidine kinase domain at the C terminus (Fig. 1A). Comparative analysis of the globin domain sequence with those of other globin proteins suggested that the N-terminal domain binds heme, similar to myoglobin, YddV, HemAT, AvGReg, BpeGReg, GsGCS, and HemDGC (Fig. 1B) (25–36). The globin domain of this GcHK displays homology with that of SWMb (34% similarity and 16% identity) and HemAT-Bs (39% similarity and 24% identity). Based on the data, we speculated that GcHK from Anaeromyxobacter sp. Fw109-5 (abbreviated as AfGcHK) is a heme-bound, globin-coupled oxygen sensor enzyme with histidine kinase activity. In this study we cloned, overexpressed, and purified the full-length AfGcHK protein and characterized its catalytic and physicochemical properties for the first time.
Oligomerization State
Gel filtration chromatographic patterns indicated that purified AfGcHK exists as a dimer in solution (supplemental Fig. S2). Dimerization was required for catalytic activity, similar to other histidine kinase proteins, which generally autophosphorylate in trans (1–4).
Optical Absorption Spectra of Purified Full-length AfGcHK
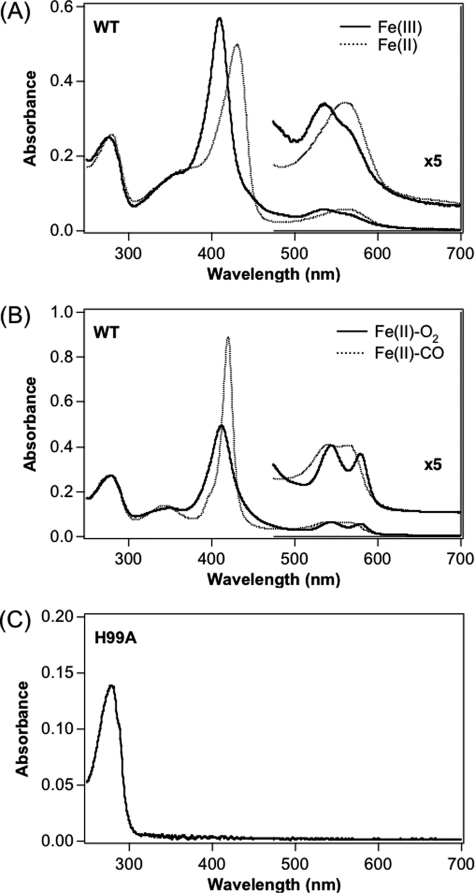
We obtained purified full-length AfGcHK containing heme (iron-protoporphyrin IX complex) at a heme:protein stoichiometry of 1:1, as determined using the pyridine hemochromogen method (37). The Fe(III) complex of purified full-length AfGcHK contained a Soret band at 411 nm and visible band at 538 nm, suggestive of a 6-coordinated low spin complex (37, 45) (Fig. 2A, Table 1). The spectrum of the Fe(III) complex was stable and did not alter over a pH range of 7.0–10.0 (data not shown), although precipitates formed below pH 6.0 (pI = 6.32). The pKa value of the Tyr-OH is 10.9, whereas the corresponding values for water associated with SWMb and human hemoglobin are 8.99 and 8.05, respectively (37). Based on the pH-insensitive spectral behavior of AfGcHK between pH 7.0 and 10.0, we suggest that the hydroxide is the axial ligand in the distal heme side, similar to EcDOS (20, 22, 43). The Fe(II) complex was a 5-coordinated high spin complex, whereas Fe(II)-O2- and Fe(II)-CO-bound proteins were 6-coordinated low spin complexes (Fig. 2B), similar to myoglobin (37, 45). Notably, the Fe(II)-O2 complex was very stable, and the spectrum of this complex did not change, even after more than 3 days at room temperature.
FIGURE 2.
Absorption spectra of the Fe(III) (solid line), Fe(II) (dotted line) (A), Fe(II)-O2 (solid line), and Fe(II)-CO (dotted line) (B) complexes of wild-type and H99A mutant (C) of full-length AfGcHK. Protein concentration was 5 μm, and buffer was 50 mm Tris-HCl, pH 8.0. Spectra of other Tyr-45 mutants and H183A mutant full-length AfGcHK were essentially similar to those of the corresponding complexes of the wild-type protein, as summarized in Table 1.
TABLE 1.
Absorption spectra of the Fe(III), Fe(II), Fe(II)-O2, and Fe(II)-CO complexes of wild-type and mutant AfGcHK
Corresponding spectra of other heme-based oxygen sensor enzymes and SWMb are shown as the reference. Proposed coordination structures are presented in parentheses. 6cLS, 6-coordinated low spin; 5cHS, 5-coordinated high spin; 6cHS, 6-coordinated high spin.
| Proteins | Fe(III) | Fe(II) | Fe(II)-O2 | Fe(II)-CO |
|---|---|---|---|---|
| AfGcHK | ||||
| WT | 411, 538 (6cLS) | 431, 559 (5cHS) | 413, 545, 580 (6cLS) | 420, 541, 565 (6cLS) |
| Y45F | 411, 535 (6cLS) | 430, 555 (5cHS) | 414, 546, 579 (6cLS) | 420, 541, 563 (6cLS) |
| Y45L | 410, 534 (6cLS) | 431, 564 (5cHS) | 415, 548, 578 (6cLS) | 421, 541, 565 (6cLS) |
| Y45W | 410, 534 (6cLS) | 431, 558 (5cHS) | 414, 547, 580 (6cLS) | 420, 543, 565 (6cLS) |
| H99A | No heme absorption | |||
| H183A | 411, 543 (6cLS) | 432, 561 (5cHS) | 413, 545, 580 (6cLS) | 421, 541, 565 (6cLS) |
| YddVa | 394, 506, 651 (5cHS) | 432, 560 (5cHS) | 413, 542, 578 (6cLS) | 420, 539, 566 (6cLS) |
| HemAT-Bsb | 402, 505, 640 (6cHS) | 431, 563 (5cHS) | 414, 543, 578 (6cLS) | 422, 543, 567 (6cLS) |
| SWMbc | 410, 505, 635 (6cHS) | 434, 556 (5cHS) | 418, 543, 581 (6cLS) | 423, 542, 579 (6cLS) |
| EcDOSd | 417, 530, 562 (6cLS) | 428, 532, 563 (6cLS) | 417, 542, 578 (6cLS) | 424, 542, 578 (6cLS) |
| BjFixLe | 395, 509, 645 (5cHS) | 437, 556 (5cHS) | 419, 545, 562 (6cLS) | 427, 548, 560 (6cLS) |
Catalytic Activities of Purified Full-length AfGcHK
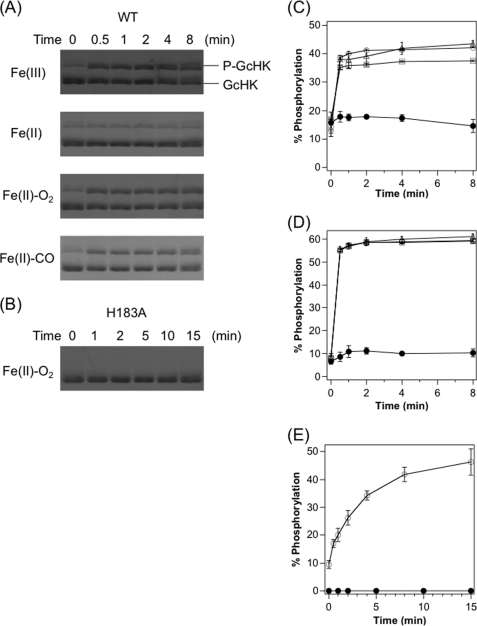
We examined the autophosphorylation activities of various heme complexes of AfGcHK using Phos-tag SDS-PAGE, which differentiates between non-phosphorylated and phosphorylated proteins (38, 39). The Fe(III), Fe(II)-O2, and Fe(II)-CO complexes clearly displayed autophosphorylation activity but not the Fe(II) complex (Fig. 3, A and C). This trend of redox- and ligand-regulated catalytic activity is similar to that observed for YddV, a globin-coupled diguanylate cyclase (28). The turnover rate of the reaction was estimated as more than 20 min−1. However, evaluation of the correct initial velocity of the catalytic reaction was not feasible because of its rapidity and completion within 5 min. In addition, an amount of autophosphorylated protein in fresh preparations of the enzyme was observed before the reaction was initiated.
FIGURE 3.
Autophosphorylation activities of full-length AfGcHKs. Phos-tag SDS-PAGE gel patterns demonstrate a time-dependent increase in phosphorylated AfGcHK (upper) and simultaneous decrease in phospho-free AfGcHK (lower) catalyzed by the various complexes of wild-type (A) and H183A (B) proteins. The H183A protein was not phosphorylated, suggesting that His-183 is the autophosphorylation site. Data were obtained at the indicated times after initiation of the reaction. Shown are time-courses for autophosphorylation of wild-type (C) and Y45F (D) full-length AfGcHKs for Fe(III) (open triangles), Fe(II) (closed circles), Fe(II)-O2 (open circles), and Fe(II)-CO (open squares) complexes. Time-courses for autophosphorylation of H99A (heme-free form) (open circles) and H183A (Fe(II)-O2 complex) (closed circles) mutants of full-length AfGcHKs are shown in E. See “Experimental Procedures” for details.
Autophosphorylation Site
To identify the autophosphorylation site, amino acid sequences in the kinase domain of GcHKs were compared with those of other orthologs of GcHK and FixL (Fig. 1C). His-183 was well conserved throughout the GcHK and FixL sequences. Accordingly, a H183A mutant was generated to examine the role of this residue in autophosphorylation activity. Fe(III) and Fe(II)-O2 complexes (active forms of wild type) of H183A lost autophosphorylation activity compared with wild-type enzyme (Fig. 3, B and E), clearly suggesting that His-183 is the autophosphorylation site in AfGcHK.
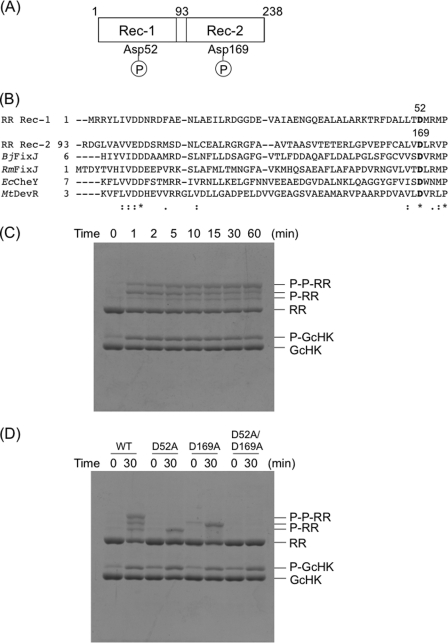
Identification of Cognate RR and Phosphotransfer Reaction from AfGcHK to RR
Anae109_2438 is located adjacent to the CheY-like RR, Anae109_2439, predicted as its cognate RR, composed of two receiver domains (Fig. 4A) (1, 2, 5–7). Purified glutathione S-transferase-tagged RR (GST-RR) was incubated with the Fe(II)-O2 complex (active form for autophosphorylation) of AfGcHK, and catalysis was initiated by adding ATP. As expected, the phosphotransfer reaction from autophosphorylated AfGcHK to GST-RR was observed as early as at 5 min after ATP addition, and GST-RR was diphosphorylated, as observed from Phos-tag SDS-PAGE (Fig. 4C). Specifically, a time-dependent decrease in phosphorylated AfGcHK (P-GcHK) was accompanied by simultaneous increase in diphosphorylated RR (P-P-RR). Interestingly, two species of monophosphorylated RR (P-RR) were also observed separately, which decreased time-dependently with a concomitant increase in the diphosphorylated species. Based on sequence analysis, Asp-52 and Asp-169 were predicted as the phosphorylation sites (Fig. 4B). Each of the two mono-phosphotransfer reactions was observed for the two single mutant proteins (D52A, D169A), supporting the proposal that Asp-52 and Asp-169 are the phosphorylation sites in RR. Moreover, the extent of protein migration of Asp-52-phosphorylated RR was smaller than that of Asp-169-phosphorylated RR on Phos-tag SDS-PAGE (Fig. 4D). The di-phosphotransfer reaction was not observed for the two single Asp-52 and Asp-169 mutants and the double Asp-52/Asp-169 mutant (Fig. 4D). Similar results were obtained with the Fe(III) complex of AfGcHK.
FIGURE 4.
The domain structure (A) and amino acid sequences of the receiver domain (B) of RR are shown. Asp-52 and Asp-169 are the assumed phosphorylation sites. C, phospho-transfer reactions to RR catalyzed by the Fe(II)-O2 complex of full-length AfGcHK are shown. Phos-tag SDS-PAGE gel patterns showing a time-dependent decrease in phosphorylated AfGcHK (P-GcHK) and simultaneous increase in diphosphorylated wild-type RR (P-P-RR) are shown. The two monophosphorylated wild-type RR bands (P-RR) increased at 5 min after initiation of the reaction but subsequently decreased in a time-dependent manner. Data were obtained at 0, 1, 2, 5, 10, 15, 30, and 60 min after initiation of the reaction. D, phospho-transfer reactions to Asp mutants of RR by the Fe(II)-O2 complex of full-length AfGcHK is shown. The upper and lower bands represent the two singly phosphorylated RR bands were abolished in the D52A and D169A mutants of RR, suggesting that these bands correspond to phosphorylated Asp-52 and Asp-169, respectively. The D52A/D169A double mutant was not phosphorylated using the same procedure. See “Experimental Procedures” for experimental details. FixJ, a response regulator for B. japonicum and R. meliloti; EcCheY, a response regulator for chemotactic signal transduction from E. coli; MtDevR, a response regulator for a two-component system and a target of phosphorylation by DevS of M. tuberculosis.
O2 Association and Dissociation Rate Constants of Wild-type Full-length AfGcHK
Because AfGcHK appears to be a heme-based oxygen sensor enzyme, we examined the O2 association and dissociation rate constants of the wild-type full-length protein. Pulse radiolysis experiments involve the instantaneous generation of hydrated electrons (eaq−), which in turn reduce the heme iron of hemoproteins and are employed to evaluate O2 association rate constants (supplemental Fig. S3) (28, 40–42). The second-order rate constant for O2 association with full-length AfGcHK was composed of a two phases with rate constants of 1.3 and 0.15 μm−1s−1 at a ∼1:1 ratio (Table 2). The O2 association rate constants (1.3 and 0.15 μm−1s−1) of AfGcHK were similar to that (0.9 μm−1s−1) of YddV (28) but markedly higher than those (0.0019–0.14 μm−1s−1) of heme-bound PAS proteins (EcDOS and BjFixL) (42, 43, 46) and lower than those (7–32 μm−1s−1) of globin proteins (SWMb, HemAT-Bs, and BpeGReg) (31, 34, 45). The rate constant for O2 dissociation, determined using the stopped-flow method, was evaluated as 0.10 s−1 and was composed of a single phase (Table 2; supplemental Fig. S4). The O2 dissociation rate constant (0.10 s−1) of AfGcHK was lower than those (4.5–23 s−1) of BjFixL (46) and the globin proteins (SWMb, YddV, HemAT-Bs, and BpeGReg) (28, 31, 34, 45) but comparable with those (0.64 s−1) of EcDOS proteins (43). The equilibrium dissociation constants of AfGcHK calculated from these rate constants were 0.077 and 0.67 μm, which were lower than those (0.64–14 μm) of other globin proteins (28, 31, 34, 45), suggesting that the novel histidine kinase has unusually high oxygen affinity as GCS.
TABLE 2.
O2 association parameters of wild-type and mutant full-length AfGcHK
Parameters of other heme-based oxygen sensor enzymes and SWMb are additionally described for reference. Experiments were repeated at least three times, and averaged values are described. Experimental errors were less than 20%. Kd values were calculated from kon and koff values.
| Proteins | kon | koff | Kd | References | |
|---|---|---|---|---|---|
| μm−1s−1 | s−1 | μm | |||
| AfGcHK | WT | 1.3, 0.15 | 0.10 | 0.077, 0.67 | This work |
| Y45F | 1.8, 0.15 | 0.35 | 0.19, 2.3 | This work | |
| Y45L | 1.5, 0.078 | 8.0 | 5.3, 100 | This work | |
| Y45W | 1.3, 0.12 | 3.4 | 2.6, 28 | This work | |
| H183A | 1.5, 0.15 | 0.11 | 0.073, 0.73 | This work | |
| YddV | 0.9 | 13 | 14 | Ref. 28 | |
| HemAT-Bs | 32 | 23 | 0.72 | Ref. 30 | |
| BpeGReg | 7.0 | 4.5 | 0.64 | Ref. 34 | |
| SWMb | 17 | 15 | 0.88 | Ref. 45 | |
| EcDOS | 0.0019 | 0.64 | 340 | Ref. 43 | |
| BjFixL | 0.14 | 20 | 140 | Ref. 46 |
CO Association and Dissociation Rate Constants of Wild-type Full-length AfGcHK
AfGcHK appears to be an oxygen sensor rather than a CO sensor, as extremely low concentrations of CO exist in cells. Nevertheless, knowledge of the CO binding kinetics is essential to clarify the molecular mechanism of AfGcHK (37, 45). The CO association and dissociation curves of the full-length enzyme were both composed of a single phase with rate constants of 0.052 μm−1s−1 and 0.0042 s−1, respectively (supplemental Figs. S5 and S6) (Table 3). The association rate constant of CO for AfGcHK was higher than those (0.00081–0.005 μm−1s−1) of heme-bound PAS proteins (EcDOS and BjFixL) (40, 43, 46) but lower than those (0.22–1.0 μm−1s−1) of the globin proteins (YddV, HemAT-Bs, BpeGReg, and SWMb) (28, 31, 34, 45). The dissociation rate constant of CO for AfGcHK was significantly lower than those (0.019–0.067 s−1) of the other oxygen sensor enzymes and SWMb (45). The equilibrium dissociation constant (0.081 μm) of CO, calculated from the association and dissociation rate constants of CO for AfGcHK, was similar to those (0.037–0.20 μm for: YddV, HemAT-Bs, BpeGReg, and SWMb) of the globin proteins (28, 31, 33, 45) but substantially lower than those of heme-bound PAS proteins (3.1–9.0 μm for EcDOS and BjFixL) (42, 43, 46).
TABLE 3.
CO association parameters of wild-type and mutant full-length AfGcHK
Parameters of other heme-based oxygen sensor enzymes and SWMb are additionally described for reference. Experiments were repeated at least three times, and averaged values are described. Experimental errors were less than 20%. Kd values were calculated from kon and koff values.
| Proteins | kon | koff | Kd | References | |
|---|---|---|---|---|---|
| μm−1s−1 | s−1 | μm | |||
| AfGcHK | WT | 0.052 | 0.0042 | 0.081 | This work |
| Y45F | 0.053 | 0.0010 | 0.019 | This work | |
| Y45L | 0.033 | 0.0042 | 0.13 | This work | |
| Y45W | 0.10 | 0.0018 | 0.018 | This work | |
| H183A | 0.042 | 0.0042 | 0.10 | This work | |
| YddV | 0.22 | 0.021 | 0.095 | Ref. 28 | |
| HemAT-Bs | 0.34 | 0.067 | 0.20 | Ref. 31 | |
| BpeGReg | 1.0 | 0.056 | 0.055 | Ref. 34 | |
| SWMb | 0.51 | 0.019 | 0.037 | Ref. 45 | |
| EcDOS | 0.00081 | 0.025 | 3.1 | Ref. 43 | |
| BjFixL | 0.005 | 0.045 | 9.0 | Ref. 46 |
Resonance Raman Spectra of Wild-type Full-length AfGcHK
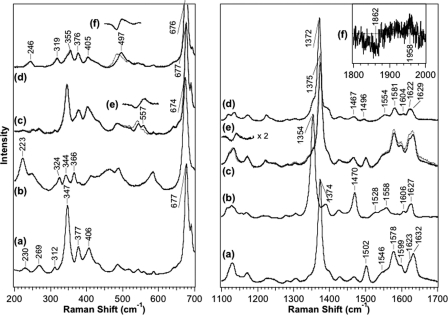
Resonance Raman spectroscopy provides useful information about the heme coordination structure and heme environment (12, 47–49). To elucidate the heme coordination structure, we obtained resonance Raman spectra of the Fe(III), Fe(II), Fe(II)-O2, and Fe(II)-CO complexes of wild-type full-length AfGcHK proteins (Fig. 5). The high frequencies of marker bands representing spin state and coordination number are summarized in supplemental Table S2 (12, 47–49). The coordination states of the heme iron complex based on Raman shifts (presented in the right-hand column in supplemental Table S2) are consistent with those suggested from optical absorption spectra (Table 1).
FIGURE 5.
Shown are resonance Raman spectra of the Fe(III) (a), Fe(II) (b), Fe(II)-O2 (c), and Fe(II)-CO (d) complexes of wild-type full-length AfGcHK in the low (left panel) and high frequency regions (right panel). The excitation wavelength was set as 413.1 nm. Band frequencies are summarized in supplemental Table S2. Resonance Raman spectra representing the Fe(II)-18O2 and Fe(II)- 13C18O complexes of wild-type full-length AfGcHK are described by the dotted lines. Difference resonance Raman spectra representing the differences (16O2-18O2) (e) in the Fe(II)-O2 complexes and differences (12C16O - 13C18O) (f) in the Fe(II)-CO complexes of full-length wild-type AfGcHK are depicted in the insets and supplemental Figs. S7–S9. See “Experimental Procedures” for experimental details.
Difference resonance Raman spectra in the low and high frequency regions for 16O2–18O2 of the Fe(II)-O2 complex and 12C16O–13C18O of the Fe(II)-CO complex of wild-type full-length AfGcHK proteins were obtained to assign the respective frequencies (Fig. 5 and supplemental Figs. S7–S9). Based on the isotope-sensitive shifts of frequencies, bands at 557, 1141, 497, and 1958 cm−1 were assigned as νFe-O2, νO-O, νFe-CO, and νC-O, respectively. Table 4 summarizes the lower and higher frequencies of the stretching modes of the Fe(II)-O2 and Fe(II)-CO complexes assigned from isotope-sensitive frequencies. The νFe-O2 frequency (557 cm−1) was similar to that (560 cm−1) of MtHb, but lower than that (569 cm−1) of SWMb, suggesting that the O2 molecule bound to the Fe(II) heme in AfGcHK is in a polar environment formed by hydrogen bond networks. The νFe-O2 frequency (557 cm−1) of AfGcHK was also lower than those (565–566 cm−1) of other globin-coupled oxygen sensors, including YddV-heme, HemAT-Bs, HemDGC, and Ascaris Hb.
TABLE 4.
Resonance Raman spectral parameters of wild-type and Tyr mutant GCS proteins, AfGcHK, YddV, HemAT-Bs, and other hemoproteins
| Proteins | νFe-His | νFe-O2 | νO-O | νFe-CO | νC-O | References | |
|---|---|---|---|---|---|---|---|
| AfGcHK | WT | 223 | 557 | 1141 | 497 | 1958 | This work |
| Y45F | 223 | 559 | 1149 | 505 | 1953 | This work | |
| YddV-heme | WT | 227 | 565 | 495 | 1965 | Ref. 28 | |
| Y43F | 227 | 559 | 505 | 1959 | Ref. 28 | ||
| HemAT-Bs | WT | 225 | 566 | 494 | 1964 | Ref. 30 | |
| Y70F | 566 | 494 | 1961 | Ref. 58 | |||
| HemDGC | WT | 226 | 566 | 1138 | 506 | 1944 | Ref. 36 |
| Y55F | 506 | Ref. 36 | |||||
| SWMb | 220 | 569 | 507 | 1947 | Refs. 47, 59, and 60 | ||
| MtHb | 226 | 560 | 500, 535 | 1960, 1916 | Refs. 50 and 51 | ||
| PcHba | 220 | 563 | 493 | 1974 | Ref. 52 | ||
| Ascaris Hb | 201 | 566 | 515, 543 | 1909, 1948 | Refs. 48 and 57 | ||
| EcDOS | 214 | 561 | 486 | 1973 | Ref. 55 | ||
| RmFixL | 211 | 571 | 498 | 1962 | Refs. 12 and 56 |
a PcHb, Paramecium caudatum hemoglobin.
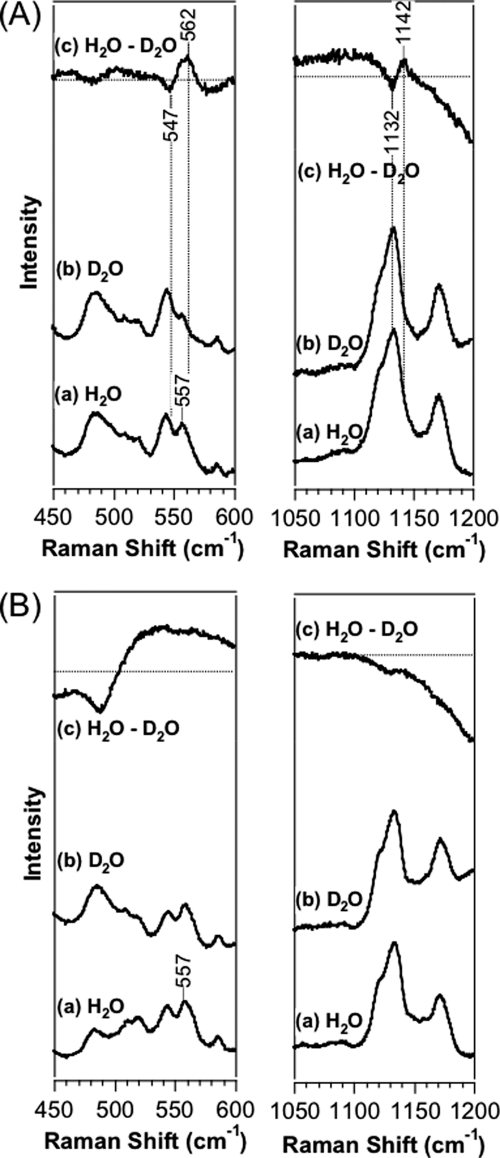
We obtained a resonance Raman spectrum of the Fe(II)-O2 complex in the deuterium solvent to determine which oxygen atom (proximal, thus interacting with Fe(II); or distal, thus not interacting with Fe(II)) of the O2 molecule interacts with the Tyr-45 OH group. Shifts in the νFe-O2 and νO-O frequencies were observed for the wild-type protein in deuterium solvent (Fig. 6A). The νFe-His frequency (223 cm−1) of AfGcHK was similar to those (225–226 cm−1) of HemAT-Bs and MtHb (30, 50, 51).
FIGURE 6.
Difference resonance Raman spectra representing the differences (16O2/H2O − 16O2/D2O) in the Fe(II)-O2 complexes of wild-type (A) and Y45F (B) full-length AfGcHK in the low (left panel) and high (right panel) frequency regions, with excitation at 413.1 nm. See “Experimental Procedures” for details.
An inverse correlation plot between νFe-CO and νC-O frequencies of the Fe(II)-CO complex (supplemental Fig. S8C) suggested that a proximal ligand trans to CO is histidine imidazole, and the CO molecule bound to the Fe(II) complex in AfGcHK is located in a hydrophobic environment (12, 49). The νFe-CO frequency (497 cm−1) of AfGcHK was close to those (493–495 cm−1) of YddV, HemAT-Bs, and P. caudatum hemoglobin (25, 27, 52), whereas the νC-O frequency (1958 cm−1) was comparable to that (1960 cm−1) of MtHb (50, 51).
Effects of Mutations at His-99 and Tyr-45 on the Heme Environment of Full-length AfGcHK
Based on the structure of the heme environment proposed from the amino acid sequences and structures determined for GCSs to date (25, 26, 32) (Fig. 1), we hypothesized that His-99 is a heme axial ligand at the proximal side and Tyr-45 is located at the heme distal side and critical for recognition or binding of the O2 molecule. Accordingly, we generated mutants at His-99 and Tyr-45 and examined the roles of these residues in spectroscopic, kinetic, and catalytic activities of the enzyme.
Optical Absorption Spectra and Kinetic Properties of Mutant Proteins of Full-length AfGcHK
Optical absorption spectroscopy studies revealed loss of heme binding ability for the H99A mutant (Fig. 2C, Table 1), supporting the possibility of His-99 as an axial ligand on the proximal side, in accordance with our proposal (Fig. 1). However, optical absorption spectra of the three Tyr-45 mutants were similar to those of wild type (Table 1). Interestingly, Y45L and Y45W mutations led to markedly enhanced O2 dissociation rate constants but did not alter the O2 association rate constants (Table 2). Thus, Tyr-45 is possibly critical in the stability of the Fe(II)-O2 complex but does not appear to be located at the O2 access channel and, therefore, may not be involved in O2 binding to the Fe(II) complex of AfGcHK. The extremely slow autooxidation rate constant of AfGcHK was not significantly altered by the mutations at Tyr-45.
The Y45F and Y45W mutations led to a 4-fold lower dissociation rate constant for CO than the wild-type protein (Table 3). However, the effects of the Tyr-45 mutations on the CO dissociation rate constant were considerably lower than those on the O2 dissociation rate constant, where up to an 80-fold increase was observed (Table 2). These findings suggest that Tyr-45 is not important for the stability of CO binding to the Fe(II) complex, distinct from the O2 molecule. The H183A mutation did not significantly alter the O2 and CO binding kinetic values (Tables 2 and 3), which appears a reasonable finding, as His-183 is the autophosphorylation site and should thus be distinct from the heme-binding domain.
Catalytic Activities of Full-length Mutant AfGcHKs
Interestingly, H99A displayed catalytic activity comparable with that of the Fe(II)-O2-complexed wild-type protein (Fig. 3E). This finding is consistent with results obtained for other oxygen sensor enzymes, YddV and EcDOS, where the heme-free forms display similar activities to the O2-bound catalytically active forms (28, 53), leading to the proposal that the protein is furnished with the heme iron complex to suppress catalysis. However, careful examination revealed that phosphorylation of H99A reached maximum levels only at 15 min after initiation of the reaction (Fig. 3E), in contrast to wild-type, where maximum phosphorylation was observed within 1 min after initiation of the reaction (Fig. 3C). The initial rate of the H99A mutant reaction appeared ∼5-fold lower than that of its wild-type counterpart. Furthermore, Tyr-45 mutants displayed catalytic activities comparable to those of wild type (activities of Y45F are shown in Fig. 3D; Y45L and Y45W showed similar activities as wild-type protein).
Resonance Raman Spectra of Full-length Mutant AfGcHKs
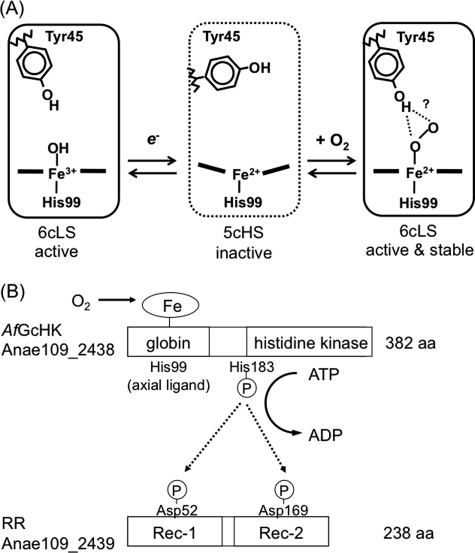
The frequencies of the Fe-O2, O-O, Fe-CO, and C-O stretching modes of Y45F are summarized in Table 4 along with frequencies of Fe-His (see supplemental Fig. S9 for difference resonance Raman spectra in the low and high frequency regions for 16O2–18O2 of the Fe(II)-O2 complex and 12C16O–13C18O of the Fe(II)-CO complex of wild-type and Y45F full-length AfGcHK proteins). The νFe-O2 (557 cm−1), νO-O (1141 cm−1), νFe-CO (497 cm−1), and νC-O (1958 cm−1) values of wild-type protein were shifted to 559, 1149, 505, and 1953 cm−1, respectively, in spectra of the Y45F mutant, implying that both O2 and CO molecules interact with the tyrosine residue. Importantly, the shifts in the νFe-O2 and νO-O frequencies observed for the wild-type protein in deuterium solvent were not detected for the Y45F mutant (Fig. 6B). Therefore, the OH group of Tyr-45 appears to form a direct hydrogen bond with the O2 molecule at the heme distal side of AfGcHK (Fig. 7A).
FIGURE 7.
A, shown is the proposed heme coordination structures relevant to catalytic activities dependent on the coordination structures and heme redox states of AfGcHK. The low spin complexes (Fe(III), Fe(II)-O2, Fe(II)-CO) are active, whereas the high spin complex (Fe(II)) is inactive. Tyr-45 OH interacts with the proximal O atom in the Fe(II)-O2 complex, but interactions with the distal O atom cannot be totally ruled out. B, shown is a proposed mechanism of globin-coupled heme-based oxygen sensing of a histidine kinase, AfGcHK, in the two-component signal transduction system. His-99 is the heme axial ligand at the proximal side of AfGcHK. The inactive high spin Fe(II) complex is activated by association of O2, forming the active low spin Fe(II)-O2 complex or autooxidized to the active low spin Fe(III) complex. His-183 is autophosphorylated by the active form of AfGcHK, and the phosphate group of phosphorylated AfGcHK is transferred to the Asp-52 and Asp-169 sites in RR. aa, amino acids; 6cLS, 6-coordinated low spin; 5cHS, 5-coordinated high spin.
DISCUSSION
Several heme-regulated oxygen or NO sensor histidine kinases have been identified, but no GcHKs are known at present (supplemental Fig. S10). In this study we have identified AfGcHK from Anaeromyxobacter sp. strain Fw109-5 and examined its catalytic, kinetic, and spectral properties for the first time.
Tyr-45 as the Axial Ligand for the Fe(III) Heme Complex
We suggest that a hydroxide group serves as an axial ligand at the heme-distal side of the six-coordinated low spin Fe(III) complex of AfGcHK (Fig. 7). Direct coordination of the Tyr-45-OH to Fe(III) heme is unlikely because the broad band (600–700 nm) characteristic of the six-coordinated high spin Fe(III) complex of the F113Y mutant of EcDOS and the H64Y mutant of SWMb (45, 53) is not observed when wild-type AfGcHK is studied (Fig. 2). However, small differences in optical absorption (Table 1) and resonance Raman spectral traces (supplemental Table S1) between the wild-type enzyme and the Tyr-45 mutant are evident. Probably, indirect interaction of the Tyr-45-OH and the Fe(III) heme complex via a hydroxide moiety (the direct axial ligand of the Fe(III) complex) or another electrostatic interaction may contribute to spectral changes associated with the Tyr-45 mutation. In Chlamydomonas chloroplast Hb (in which Tyr-63-OH serves as the direct axial ligand of the Fe(III) heme complex), an H/D-sensitive band at 502 cm−1 (assigned to the Fe-O[Tyr] stretching mode) was observed, and Tyr-63 mutation caused significant shifts in the resonance Raman spectral patterns (66). In contrast, the resonance Raman spectral properties characteristic of direct Tyr-OH ligation were not observed when AfGcHK was studied. We further discuss the resonance Raman spectral characteristics of AfGcHK later in this section.
Autophosphorylation
Phos-tag SDS-PAGE patterns (Fig. 3) demonstrating autophosphorylation activities indicated termination of the reactions at around 50–60% phosphorylation. These phenomena may occur due to equilibrium between the autophosphorylation and phosphatase reactions. Because GcHK is about 50% phosphorylated in general and other histidine kinases are only 5–10% phosphorylated at the final stage, the Phos-tag SDS-PAGE patterns observed for AfGcHK are not surprising. It should be emphasized that the Phos-tag SDS-PAGE method is superior to the radioisotope method using [32P]ATP to evaluate phosphorylated protein with the kinase, as it can effectively detect trace amounts of phosphorylated protein in the fresh protein preparation. No pre-autophosphorylated protein was detected for the inactive H183A mutant (Fig. 3, B and E), validating the superiority of Phos-tag SDS-PAGE over the radioisotope method (1–4, 19).
Mechanism of Catalytic Activation
For AfGcHK, all the active species are low spin complexes (Fe(III), Fe(II)-O2, and Fe(II)-CO complexes), whereas the high spin complex (Fe(II) complex) is inactive. In crystal structures of myoglobin and other heme proteins, the iron atom projects out 0.4–0.6 Å from the porphyrin plane in the Fe(II) high spin complex but is on the porphyrin plane in the low spin Fe(II)-O2 complex (37, 45, 61, 62). Therefore, the Fe-His bond length on the proximal side appears crucial for catalytic activation of globin-coupled sensors.
The initial reaction rate of the H99A protein (axial ligand mutant; heme-free form) appeared 5-fold lower than that of wild-type protein (Fig. 3, C and E). Autophosphorylation is exerted in a crosswise manner in that kinase on one subunit phosphorylates His-183 on another subunit. It is speculated that Ala-99 interacts directly or indirectly with the kinase active site on another subunit and partially hampers catalysis in the heme-free H99A mutant. Heme binding to His-99 as the proximal axial ligand of the wild-type protein may assist in regulation of the catalytic reaction with the aid of heme redox changes and/or external ligand (O2, CO) binding to the heme iron.
On the other hand, distal Tyr-45 may be critical for oxygen sensing. An important role of the Tyr residue in the heme distal side of oxygen sensor proteins with the globin-, GAF- and H-NOX domains in oxygen sensing has been documented (18, 28, 32, 36, 54).
The autophosphorylated amounts (∼60%) of Y45F (Fig. 3D) appeared significantly higher than those (∼40%) of wild-type protein (Fig. 3C). Tyr-45 may interfere with catalysis in a negative manner, as suggested for His-99. Because the reaction was so rapid and almost completed within 1 min using the present method, correct catalytic parameters, such as Vmax and Km values, could not be obtained.
Stability of the Fe(II)-O2 Complex and O2 and CO Binding Kinetics
One of the characteristic features of AfGcHK is the high stability of the Fe(II)-O2 complex. Our findings suggest that AfGcHK is constitutively active as the Fe(II)-O2 (or Fe(III)) complex under aerobic conditions but inactive as the Fe(II) complex under hypoxia or anaerobic conditions. The O2 affinity (Kd 0.077 and 0.67 μm) of AfGcHK is the highest among the known oxygen sensor enzymes and SWMb (Table 2) but differs from those (EcDOS, 340 μm; BjFixL, 140 μm) of heme-bound PAS oxygen sensor enzymes (42, 43, 46). AfGcHK may need to detect the presence of trace amounts of oxygen in the bacterial anaerobic environment to switch catalysis, whereas PAS oxygen sensor enzymes are required to detect lowering of the oxygen concentration under normoxia conditions (260 μm or saturated O2 concentration, 1.3 mm) in an aerobic bacterial environment.
Effects of Tyr-45 mutations on the koff value were more marked than those on the kon value for O2 binding (Table 2). These findings are in accordance with the resonance Raman spectral findings in that Tyr-45 mutations shifted the frequencies of the Fe(II)-O2 complex (Table 4). A koff value can be used to evaluate ligand dissociation from a ligand-heme complex and thus reflects the heme-distal structure; this is the region in which a ligand becomes dissociated (37, 45). However, a kon value can be used to evaluate the characteristics of a ligand-access channel (37, 45). Therefore, it is suggested that the Tyr-45 side chain interacts with the O2 molecule bound to Fe(II) heme by forming a hydrogen bond network on the heme distal side. Note that the koff value of ligand dissociation for the heme complex correlated with resonance Raman frequencies, as shown below. Mutations at the distal Tyr residue of MtHb enhanced the koff value of O2 by 100-fold, consistent with the results for AfGcHK (50, 51).
Resonance Raman Spectra
The iron-histidine bond-stretching mode (νFe-His) is only observed in the 5-coordinate Fe(II) complex and is very sensitive to the protein matrix surrounding the histidine residue (48). Because the frequency of this mode of AfGcHK (223 cm−1) is similar to those of other heme proteins, such as SWMb (220 cm−1), HemAT-Bs (225 cm−1), and MtHb (226 cm−1), it is suggested that the Nϵ atom of the proximal histidine of AfGcHK interacts only weakly with a nearby polar side chain, distinct from horseradish peroxidase (244 cm−1). The hydrogen bond of the proximal histidine of the peroxidase enzyme appears to be important in terms of stabilization of reactive intermediate oxygen species including Compound I. The frequency differences between SWMb (220 cm−1) and those of GCS proteins/truncated globin (AfGcHK, 223 cm−1; HemAT-Bs, 225 cm−1; MtHb, 226 cm−1) may be attributed to variation in the strength of hydrogen bonds associated with the oxygen-transfer and gas-sensing functions of such proteins.
The νFe-O2 frequency (557 cm−1) of AfGcHK was similar to those (560 cm−1) of MtHb (50, 51). Interestingly, Tyr mutations on the heme distal side of MtHb led to a shift in the νFe-O2 frequency from 560 to 570 cm−1. This spectral behavior is analogous to that of AfGcHK, suggesting that the proximal oxygen atom of the O2 molecule bound to the Fe(II) heme forms a hydrogen bond with the Tyr-OH group (50, 51). The shifts in the νFe-O2 and νO-O frequencies of the wild-type protein in deuterium solvent were not observed for Y45F (Fig. 6B), supporting the presence of hydrogen bond(s) between the O2 molecule and Tyr-45 OH in wild-type AfGcHK. A crystal structure study of the Fe(II)-O2 complex of MtHb suggested that both oxygen atoms in the O2 molecule are almost at the same hydrogen-bonding distance from the phenolic OH group of Tyr-33 (average value, 3.12 Å) (63).
The resonance Raman spectral frequencies (νFe-CO, 497 cm−1; νC-O, 1958 cm−1) of the Fe(II)-CO complex of AfGcHK were also similar to those (νFe-CO, 500 cm−1; νC-O, 1960 cm−1) of MtHb (Table 4), suggesting comparable binding of the CO molecule to Fe(II) heme in AfGcHK to that in MtHb. The Y45F mutation led to increased νFe-CO frequency but decreased νC-O frequency of the Fe(II)-CO complex of AfGcHK. We propose that the CO molecule does not form a hydrogen bond in the wild-type protein but forms a hydrogen bond(s) with a nearby polar residue(s) or water molecule(s) in the Y45F mutant based on the inverse correlation plot between νFe-CO and νC-O (12, 49). The Y45F mutation may induce steric rearrangement of a neighboring polar amino acid residue (or the water molecule) to face and/or to interact with CO.
The Fe(III)-OH stretching frequency should be observed between 450 and 555 cm−1 (47). However, the bands of the Fe(III) complex of AfGcHK were very weak. We could not even detect clear differences between spectra obtained in H2O and D2O. Further attempts are required to probe Fe(III)-OH coordination from the resonance Raman spectral point of view.
Physiological Functions
In the absence of clear phenotypic data, it is difficult to determine the actual physiological function(s) of RR encoded by the Anae109_2439 gene. RR identified in this study has two tandem receiver domains, whereas other documented RRs consist of a single receiver domain, such as CheY, and affect protein localization, function as phosphate sinks, or control chemotactic or chemokinetic responses (5–7). The two-component systems associated with this gene have additionally been identified in Anaeromyxobacter dehalogenans and Myxococcus xanthus, among the sequenced bacteria. Anaeromyxobacter exhibits both aerobic and anaerobic growth. On the other hand, Myxococcus is an obligate aerobe and cannot survive under anaerobic conditions. Therefore, the two-component system may regulate specific physiological functions, such as fruiting body formation and sporulation, for these soil slime bacteria, which require optimal oxygen concentrations to exert adequate physiological functions for cell survival (64, 65).
Summary
In this study we have isolated and characterized the novel heme-bound globin-coupled histidine kinase, AfGcHK, from Anaeromyxobacter sp. Fw109-5 for the first time. The low spin Fe(III), Fe(II)-O2, and Fe(II)-CO complexes appear to be the active forms, and the high spin Fe(II) complex appear to be the inactive form of the enzyme. Spectral data on proteins with mutations in the heme domain suggest that His-99 is the proximal axial ligand to the heme iron complex, whereas Tyr-45 is located in the heme distal side and forms electrostatic interactions with the O2 molecule bound to the Fe(II) complex. An autophosphorylation site, His-183, on AfGcHK, and phosphorylation sites Asp-52 and Asp-169 on RR have been further identified. Histidine kinases with heme-bound sensor domains having different folds, such as PAS and GAF (supplemental Fig. S10), have been documented, but no histidine kinases with the heme-bound globin fold have been reported to date. To our knowledge, AfGcHK is a first known globin-coupled oxygen sensor histidine kinase critical for the two-component system in Anaeromyxobacter sp.
Supplementary Material
Acknowledgment
We thank Yukari Sekine at Hokkaido University for assistance with resonance Raman measurements.
This work was supported in part by the Management Expenses Grants for National Universities Corporations from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT) (to T. S.) as well as a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists (to K. Kitanishi).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S10.
- RR
- response regulator
- GcHK
- globin-coupled histidine kinase
- AfGcHK
- a GcHK from Anaeromyxobacter sp. Fw109-5
- DGC
- diguanylate cyclase
- AvGReg
- a globin-coupled oxygen-sensor enzyme with the DGC activity from Azotobacter vinelandii
- BpeGReg
- a globin-coupled oxygen-sensor enzyme with the DGC activity from Bordetella pertussis
- EcDOS
- E. coli direct oxygen sensor or heme-regulated phosphodiesterase from E. coli
- Fe(III)
- Fe(III)-protoporphyrin IX complex, Fe(III) heme complex, or hemin
- Fe(II)
- Fe(II)-protoporphyn IX complex or Fe(II) heme complex
- FixL
- an oxygen-sensor histidine kinase with the heme-bound PAS domain that regulates nitrogen fixation in R. meliloti (RmFixL) or B. japonicum (BjFixL)
- GAF
- domain conserved in cyclic GMP-specific and stimulated phosphodiesterases, adenylate cyclases, and E. coli formate hydrogen lyase transcriptional activator
- GCS
- globin-coupled oxygen sensor
- GsGCS
- GCS from Geobacter sulfurreducens
- HemAT-Bs
- a globin-coupled oxygen-sensor enzyme from Bacillus subtilis
- HemDGC
- a heme-containing DGC from Desulfotalea psychrophila
- Hb
- hemoglobin, MtHb, M. tuberculosis Hb
- PAS
- an acronym formed from Per (Drosophila period clock protein)-Arnt (vertebrate aryl hydrocarbon receptor nuclear translocator)-Sim (Drosophila single-minded protein)
- SWMb
- sperm whale myoglobin
- YddV
- heme-bound DGC from E. coli or EcDosC.
REFERENCES
- 1. Inouye M., Dutta R. Eds. (2003) Histidine Kinases in Signal Transduction, Academic Press, Inc., San Diego, CA [Google Scholar]
- 2. Krämer R., Jung K. Eds. (2010) Bacterial Signaling, Wiley-VCH, Weinheim, Germany [Google Scholar]
- 3. Casino P., Rubio V., Marina A. (2010) Curr. Opin. Struct. Biol. 20, 763–771 [DOI] [PubMed] [Google Scholar]
- 4. Cheung J., Hendrickson W. A. (2010) Curr. Opin. Microbiol. 13, 116–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gao R., Mack T. R., Stock A. M. (2007) Trends Biochem. Sci. 32, 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Galperin M. Y. (2010) Curr. Opin. Microbiol. 13, 150–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jenal U., Galperin M. Y. (2009) Curr. Opin. Microbiol. 12, 152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Green J., Crack J. C., Thomson A. J., LeBrun N. E. (2009) Curr. Opin. Microbiol. 12, 145–151 [DOI] [PubMed] [Google Scholar]
- 9. Igarashi J., Kitanishi K., Shimizu T. (2011) in Handbook of Porphyrin Science (Kadish K. M., Smith K. M., Guilard R. eds) Vol. 15, pp. 399–460, World Scientific, Hackensack, NJ [Google Scholar]
- 10. Gilles-Gonzalez M. A., Gonzalez G. (2005) J. Inorg. Biochem. 99, 1–22 [DOI] [PubMed] [Google Scholar]
- 11. Nakamura H., Kumita H., Imai K., Iizuka T., Shiro Y. (2004) Proc. Nat. Acad. Sci. U.S.A. 101, 2742–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Uchida T., Kitagawa T. (2005) Acc. Chem. Res. 38, 662–670 [DOI] [PubMed] [Google Scholar]
- 13. Gilles-Gonzalez M. A., Gonzalez G. (2004) J. Appl. Physiol. 96, 774–783 [DOI] [PubMed] [Google Scholar]
- 14. Sardiwal S., Kendall S. L., Movahedzadeh F., Rison S. C., Stoker N. G., Djordjevic S. (2005) J. Mol. Biol. 353, 929–936 [DOI] [PubMed] [Google Scholar]
- 15. Ioanoviciu A., Yukl E. T., Moënne-Loccoz P., Ortiz de Montellano P. R. (2007) Biochemistry 46, 4250–4260 [DOI] [PubMed] [Google Scholar]
- 16. Sousa E. H., Tuckerman J. R., Gonzalez G., Gilles-Gonzalez M. A. (2007) Protein Sci. 16, 1708–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kumar A., Toledo J. C., Patel R. P., Lancaster J. R., Jr., Steyn A. J. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11568–11573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boon E. M., Marletta M. A. (2005) J. Inorg. Biochem. 99, 892–902 [DOI] [PubMed] [Google Scholar]
- 19. Ito Y., Nakagawa S., Komagata A., Ikeda-Saito M., Shiro Y., Nakamura H. (2009) FEBS Lett. 583, 2244–2248 [DOI] [PubMed] [Google Scholar]
- 20. Sasakura Y., Yoshimura-Suzuki T., Kurokawa H., Shimizu T. (2006) Acc. Chem. Res. 39, 37–43 [DOI] [PubMed] [Google Scholar]
- 21. Chang A. L., Tuckerman J. R., Gonzalez G., Mayer R., Weinhouse H., Volman G., Amikam D., Benziman M., Gilles-Gonzalez M. A. (2001) Biochemistry 40, 3420–3426 [DOI] [PubMed] [Google Scholar]
- 22. Tanaka A., Takahashi H., Shimizu T. (2007) J. Biol. Chem. 282, 21301–21307 [DOI] [PubMed] [Google Scholar]
- 23. Yoshioka S., Kobayashi K., Yoshimura H., Uchida T., Kitagawa T., Aono S. (2005) Biochemistry 44, 15406–15413 [DOI] [PubMed] [Google Scholar]
- 24. Moskvin O. V., Kaplan S., Gilles-Gonzalez M. A., Gomelsky M. (2007) J. Biol. Chem. 282, 28740–28748 [DOI] [PubMed] [Google Scholar]
- 25. Freitas T. A., Hou S., Alam M. (2003) FEBS Lett. 552, 99–104 [DOI] [PubMed] [Google Scholar]
- 26. Freitas T. A., Saito J. A., Hou S., Alam M. (2005) J. Inorg. Biochem. 99, 23–33 [DOI] [PubMed] [Google Scholar]
- 27. Tuckerman J. R., Gonzalez G., Sousa E. H., Wan X., Saito J. A., Alam M., Gilles-Gonzalez M. A. (2009) Biochemistry 48, 9764–9774 [DOI] [PubMed] [Google Scholar]
- 28. Kitanishi K., Kobayashi K., Kawamura Y., Ishigami I., Ogura T., Nakajima K., Igarashi J., Tanaka A., Shimizu T. (2010) Biochemistry 49, 10381–10393 [DOI] [PubMed] [Google Scholar]
- 29. Hou S., Larsen R. W., Boudko D., Riley C. W., Karatan E., Zimmer M., Ordal G. W., Alam M. (2000) Nature 403, 540–544 [DOI] [PubMed] [Google Scholar]
- 30. Aono S., Kato T., Matsuki M., Nakajima H., Ohta T., Uchida T., Kitagawa T. (2002) J. Biol. Chem. 277, 13528–13538 [DOI] [PubMed] [Google Scholar]
- 31. Zhang W., Olson J. S., Phillips G. N., Jr. (2005) Biophys. J. 88, 2801–2814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang W., Phillips G. N., Jr. (2003) Structure 11, 1097–1110 [DOI] [PubMed] [Google Scholar]
- 33. Thijs L., Vinck E., Bolli A., Trandafir F., Wan X., Hoogewijs D., Coletta M., Fago A., Weber R. E., Van Doorslaer S., Ascenzi P., Alam M., Moens L., Dewilde S. (2007) J. Biol. Chem. 282, 37325–37340 [DOI] [PubMed] [Google Scholar]
- 34. Wan X., Tuckerman J. R., Saito J. A., Freitas T. A., Newhouse J. S., Denery J. R., Galperin M. Y., Gonzalez G., Gilles-Gonzalez M. A., Alam M. (2009) J. Mol. Biol. 388, 262–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pesce A., Thijs L., Nardini M., Desmet F., Sisinni L., Gourlay L., Bolli A., Coletta M., Van Doorslaer S., Wan X., Alam M., Ascenzi P., Moens L., Bolognesi M., Dewilde S. (2009) J. Mol. Biol. 386, 246–260 [DOI] [PubMed] [Google Scholar]
- 36. Sawai H., Yoshioka S., Uchida T., Hyodo M., Hayakawa Y., Ishimori K., Aono S. (2010) Biochim. Biophys. Acta 1804, 166–172 [DOI] [PubMed] [Google Scholar]
- 37. Antonini E., Brunori M. (1971) Hemoglobin and Myoglobin in Their Reactions with Ligands, North-Holland Publishing Co., Amsterdam, The Netherlands [Google Scholar]
- 38. Yamada S., Nakamura H., Kinoshita E., Kinoshita-Kikuta E., Koike T., Shiro Y. (2007) Anal. Biochem. 360, 160–162 [DOI] [PubMed] [Google Scholar]
- 39. Igarashi J., Murase M., Iizuka A., Pichierri F., Martinkova M., Shimizu T. (2008) J. Biol. Chem. 283, 18782–18791 [DOI] [PubMed] [Google Scholar]
- 40. Kobayashi K., Tagawa S., Daff S.., Sagami I., Shimizu T. (2001) J. Biol. Chem. 276, 39864–39871 [DOI] [PubMed] [Google Scholar]
- 41. Nakajima H., Nakagawa E., Kobayashi K., Tagawa S., Aono S. (2001) J. Biol. Chem. 276, 37895–37899 [DOI] [PubMed] [Google Scholar]
- 42. Kobayashi K., Tanaka A., Takahashi H., Igarashi J., Ishitsuka Y., Yokota N., Shimizu T. (2010) J. Biochem. 148, 693–703 [DOI] [PubMed] [Google Scholar]
- 43. Taguchi S., Matsui T., Igarashi J., Sasakura Y., Araki Y., Ito O., Sugiyama S., Sagami I., Shimizu T. (2004) J. Biol. Chem. 279, 3340–3347 [DOI] [PubMed] [Google Scholar]
- 44. Ishitsuka Y., Araki Y., Tanaka A., Igarashi J., Ito O., Shimizu T. (2008) Biochemistry 47, 8874–8884 [DOI] [PubMed] [Google Scholar]
- 45. Springer B. A., Sligar S. G., Olson J. S., Phillips. G. N., Jr. (1994) Chem. Rev. 94, 699–714 [Google Scholar]
- 46. Gilles-Gonzalez M. A., Gonzalez G., Perutz M. F., Kiger L., Marden M. C., Poyart C. (1994) Biochemistry 33, 8067–8073 [DOI] [PubMed] [Google Scholar]
- 47. Spiro T. G., Li X.-Y. (1988) in Biological Applications of Raman Spectroscopy (Spiro T. G. ed) Vol. 3, pp. 1–37, John Wiley & Sons, Inc., New York [Google Scholar]
- 48. Kitagawa T. (1988) in Biological Applications of Raman Spectroscopy (Spiro T. G. ed) Vol. 3, pp. 97–131, John Wiley & Sons, Inc., New York [Google Scholar]
- 49. Spiro T. G., Wasbotten I. H. (2005) J. Inorg. Biochem. 99, 34–44 [DOI] [PubMed] [Google Scholar]
- 50. Couture M., Yeh S. R., Wittenberg B. A., Wittenberg J. B., Ouellet Y., Rousseau D. L., Guertin M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11223–11228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yeh S. R., Couture M., Ouellet Y., Guertin M., Rousseau D. L. (2000) J. Biol. Chem. 275, 1679–1684 [DOI] [PubMed] [Google Scholar]
- 52. Das T. K., Weber R. E., Dewilde S., Wittenberg J. B., Wittenberg B. A., Yamauchi K., Van Hauwaert M. L., Moens L., Rousseau D. L. (2000) Biochemistry 39, 14330–14340 [DOI] [PubMed] [Google Scholar]
- 53. Tanaka A., Shimizu T. (2008) Biochemistry 47, 13438–13446 [DOI] [PubMed] [Google Scholar]
- 54. Yukl E. T., Ioanoviciu A., Nakano M. M., Ortiz de Montellano P. R., Moënne-Loccoz P. (2008) Biochemistry 47, 12532–12539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sato A., Sasakura Y., Sugiyama S., Sagami I., Shimizu T., Mizutani Y., Kitagawa T. (2002) J. Biol. Chem. 277, 32650–32658 [DOI] [PubMed] [Google Scholar]
- 56. Mukai M., Nakamura K., Nakamura H., Iizuka T., Shiro Y. (2000) Biochemistry 39, 13810–13816 [DOI] [PubMed] [Google Scholar]
- 57. Das T. K., Friedman J. M., Kloek A. P., Goldberg D. E., Rousseau D. L. (2000) Biochemistry 39, 837–842 [DOI] [PubMed] [Google Scholar]
- 58. Ohta T., Yoshimura H., Yoshioka S., Aono S., Kitagawa T. (2004) J. Am. Chem. Soc. 126, 15000–15001 [DOI] [PubMed] [Google Scholar]
- 59. Ramsden J., Spiro T. G. (1989) Biochemistry 28, 3125–3128 [DOI] [PubMed] [Google Scholar]
- 60. Kitagawa T., Nagai K., Tsubaki M. (1979) FEBS Lett. 104, 376–378 [DOI] [PubMed] [Google Scholar]
- 61. Vojtechovský J., Chu K., Berendzen J., Sweet R. M., Schlichting I. (1999) Biophys. J. 77, 2153–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Park S. Y., Yokoyama T., Shibayama N., Shiro Y., Tame J. R. (2006) J. Mol. Biol. 360, 690–701 [DOI] [PubMed] [Google Scholar]
- 63. Milani M., Pesce A., Ouellet Y., Ascenzi P., Guertin M., Bolognesi M. (2001) EMBO J. 20, 3902–3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shi X., Wegener-Feldbrügge S., Huntley S., Hamann N., Hedderich R., Søgaard-Andersen L. (2008) J. Bacteriol. 190, 613–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zusman D. R., Scott A. E., Yang Z., Kirby J. R. (2007) Nat. Rev. Microbiol. 5, 862–872 [DOI] [PubMed] [Google Scholar]
- 66. Das T. K., Couture M., Lee H. C., Peisach J., Rousseau D. L., Wittenberg B. A., Wittenberg J. B., Guertin M. (1999) Biochemistry 38, 15360–15368 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.