Abstract
The serine/threonine checkpoint kinase 2 (Chk2) is an attractive molecular target for the development of small molecule inhibitors to treat cancer. Here, we report the rational design of Chk2 inhibitors that target the gatekeeper-dependent hydrophobic pocket located behind the adenine-binding region of the ATP-binding site. These compounds exhibit IC50 values in the low nanomolar range and are highly selective for Chk2 over Chk1. X-ray crystallography was used to determine the structures of the inhibitors in complex with the catalytic kinase domain of Chk2 to verify their modes of binding.
Keywords: rational drug design, structure-based drug design, kinase inhibitors
1. Introduction
The serine/threonine checkpoint kinase 2 (Chk2) is an important component of the intracellular signaling network that responds to DNA damage and maintains genomic integrity[1–3]. Activation of Chk2 is mediated primarily by ATM or DNA-PK (also ATR and hMPs1) by phosphorylation of Thr68 in the SQ/TQ cluster domain [4] which initiates homodimerization of Chk2 monomers followed by trans-activating autophosphorylation of Thr383 and Thr387 [5] and subsequently the cis-phosphorylation of Ser516 [6]. The activated Chk2 monomers phosphorylate a number of downstream substrates that are involved in the regulation of the cell cycle [7], DNA repair [8,9], chromosome stability [10], and/or the initiation of apoptosis [1].
Chk2 has been identified as a potential molecular target for anti-cancer drug design, particularly in p53-defective tumors [1,11–13] and recently in DNA repair (Mus81)-deficient tumors [14]. Selective inhibition of Chk2 in p53-deficient tumor cell lines in synergy with DNA-damaging chemotherapeutics may help increase the sensitivity of these tumors to current chemotherapy agents by targeting the G2 checkpoint and thereby blocking the protective mechanisms conferred by cell-cycle checkpoints and DNA repair pathways [1,11,12,15–20]. Previous studies have shown that the Chk2 specific inhibitor, PV1019, potentiated the cytotoxicity of campothecin and topotecan in three ovarian cancer cell lines that had high levels of endogeneously activated Chk2 [21]. Another advantageous feature of selective Chk2 inhibitors may be their ability to protect normal cells against ionizing radiation and chemotherapeutics by blocking Chk2-dependent activation of p53 [1,22]. Prior studies have shown that treatment of mouse thymocytes and human T-cells with selective Chk2 inhibitors provides protection against radiation [21,23,24]. Finally, the use of selective Chk2 inhibitors by themselves may confer some therapeutic benefit since Chk2 plays important roles in tumor cell adaptation to changes resulting from the cycling nature of hypoxia and reoxygenation found in solid tumors [17], the activation of BRCA1 [8,10], and in the release of survivin [15]. Indeed, the Chk2 inhibitor PV1019 exhibited antiproliferative effects against tumor cell lines from the NCI-60 with high endogenous levels of activated Chk2 [21].
In this study, we used X-ray crystallography to assist in further modification of the Chk2 inhibitor PV1019 that was developed by our laboratories [21]. Based on prior crystallographic evidence, the binding mode of PV1019 in the ATP-binding site of Chk2 revealed that the gatekeeper-dependent hydrophobic pocket (GDHP) was partially occupied by the methyl group of PV1019. The size of the pocket is sufficient to accommodate additional substituents on the methyl group. Since the potency and selectivity of other kinase inhibitors has been improved by targeting analogous GDHPs [25,26], we sought to modify the chemical scaffold of PV1019 in order to achieve binding to this pocket in Chk2.
2. Materials and methods
2.1 Biochemical characterization of inhibitors
The catalytic domain of human Chk2 (Ser210-Glu531) was expressed and purified as described [27]. The IMAP Screening Express Kit (Molecular Devices, Sunnyvale, CA) was used for conducting inhibition assays. The compounds used in this study were synthesized by Provid Pharmaceuticals and dissolved in DMSO. Reactions were performed using recombinant human Chk2, RSK2 and Chk1 (Millipore, Billerica, MA)) with compounds in reaction buffer (10 mM Tris-HCl, pH 7.2, 10 mM magnesium chloride, 0.1% bovine serum albumin, 1 mM dithiothreitol, 10 mM ATP, and 100 nM peptide substrate) in a total volume of 5 μL in 384-well plates for 60 minutes at room temperature. Substrates used in the assay were 5FAM-AMRLERQDSIFYPK-NH2 for Chk2, 5FAM-ALKLVRYPSFVITAK-NH2 for Chk1, and 5FAM-AKRRRLSSLRA-OH for RSK2 (all from Molecular Devices). 15 μL of IMAP binding reagent were added to each well, the plates were incubated for 30 minutes at room temperature, and fluorescence polarization was measured using a Tecan Ultra plate reader at wavelengths of 485 nm for excitation and 535 nm for emission. Each screening plated contained staurosporine as a positive control.
2.2 Crystallization and Structure Solution
Crystallization of Chk2-inhibitor complexes was performed as previously described [27]. X-ray diffraction data were collected from crystals held at approximately 100 K, using a 1.0 Å wavelength, an oscillation angle of 1.0°, and a 3 second exposure time. The data were integrated and scaled using HKL3000 [28]. The structures were solved by molecular replacement with MOLREP [29] using the coordinates of the Chk2-PV1019 complex (PDB ID: 2W7X) and refined with REFMAC5 [30]. Model validation was performed with MolProbity [31]. All data collection and refinement statistics are presented in Table 2.
Table 2.
X-ray data collection and refinement statistics
| Chk2 complex | PV1322 | PV1352 | PV1162 |
| Space group | P3221 | P3221 | P3221 |
| Unit cell a=b, c (Å) | 90.6, 93.6 | 91.2, 93.5 | 91.2, 93.5 |
| Resolution (Å)a | 50–1.89 (1.97–1.89) | 50–2.10 (2.18–2.10) | 50–2.20 (2.28–2.20) |
| Total/Unique Reflections | 171881/34829 | 291871/26631 | 160173/23346 |
| Completeness (%) | 97.7 (99.4) | 99.8 (100) | 99.7 (100) |
| Redundancy | 5.0 (4.9) | 11.0 (10.2) | 6.9 (6.9) |
| I/σ(I) | 14.0 (2.4) | 53.6 (3.8) | 35.1 (3.7) |
| Rsymb | 0.088 (0.605) | 0.055 (0.675) | 0.070 (0.548) |
| Resolution | 50–1.90 | 50–2.10 | 50–2.20 |
| No. of reflections (refinement/Rfree | 33074/1753 | 25239/1338 | 22111/1200 |
| R/Rfreec | 0.199/0.234 | 0.195/0.225 | 0.196/0.226 |
| No. of atoms/Mean B factor (Å) | |||
| Protein | 2262/42.9 | 2285/47.2 | 2299/47.7 |
| Inhibitor | 28/39.0 | 30/54.9 | 29/40.2 |
| Water | 222/32.8 | 173/29.6 | 138/23.5 |
| Ion | 4/37.6 | 4/70.3 | 4/52.6 |
| PDB ID | 2YIQ | 2YIR | 2YIT |
Values in parentheses are for reflections in the highest resolution shell.
Rsym = Σhkl Σi |Ii (hkl) 〈I(hkl)〉|/Σhkl Σi Ii(hkl), where <I(hkl)> is the mean intensity of multiply recorded reflections.
R = Σ |Fobs(hkl) – Fcalc(hkl)|/Σ |Fobs(hkl)|. Rfree is the R value calculated for 5% of the data set not included in the refinement.
3. Results and Discussion
3.1 Structure of Chk2 in complex with PV1322
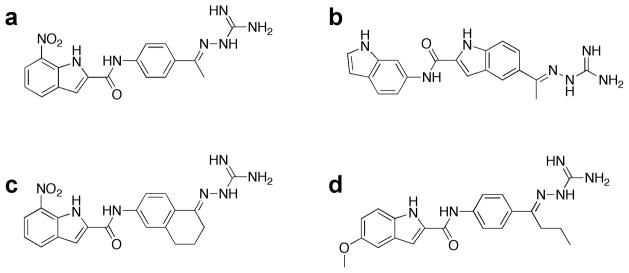
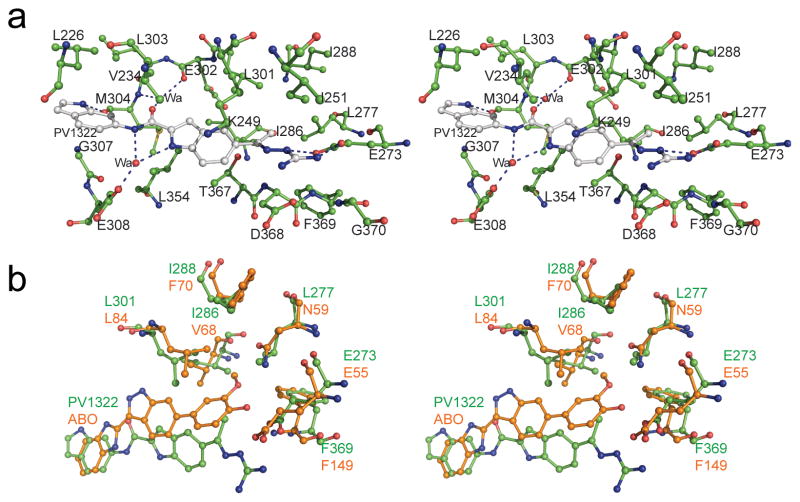
Using the crystal structure of Chk2 in complex with PV1019 as the starting point for further optimization [21], we modified the core scaffold of PV1019 (Fig. 1a) by designing the indoyl-indole analog PV1322 (Fig. 1b). In the kinase inhibition assay, PV1322 exhibited an IC50 value of 12.67 nM and was selective for Chk2 over Chk1 (IC50=34 μM) and RSK2 (IC50>100 μM). The indolyl-indole modification of PV1019 resulted in a new lead series for Chk2 by replacement of the core aryl ring of the phenyl guanidinohydrazone. The 1.89 Å resolution crystal structure of the Chk2-PV1322 complex revealed that incorporation of the indolyl-indole moiety reverses the directionality of the amide bond linker between the two aryl ring systems while at the same time retaining the water-mediated hydrogen bonds between the carbonyl oxygen and the backbone amide NH of Met304 and the backbone carbonyl oxygen of Glu302 in the hinge region of Chk2 via water 221 (Fig. 2a). The core indole group retains several of the van der Waals interactions between the aryl ring and the cluster of aliphatic residues in the ATP-binding site including Val234, Leu301, Leu354, the methyl group of Thr367, and the aliphatic portion of the Lys249 side chain. Water 57 mediates a hydrogen-bonding network involving the carboxylate side chain of Glu308, the indole NH and the nitrogen located between the carbonyl group and terminal indole. The terminal guanidine moiety of PV1322 maintains its hydrogen bonds with the Glu273 side chain, as seen in the Chk2-PV1019 complex. Replacement of the 7-nitro-indole group of PV1019 with the indole in PV1322 results in the indole binding to the hinge region via a hydrogen bond between the indole NH and the backbone carbonyl oxygen of Met304.
Fig 1.
Chemical structures of (a) PV1019, (b) PV1322, (c) PV1352 and (d) PV1162.
Fig. 2.
(a) Stereo view of PV1322 (carbon atoms in gray) in complex with Chk2 (carbon atoms in green). (b) Superimposed coordinates of the Chk2-PV1322 (green) complex with those of Chk1-ABO (orange) complex (PDB code: 2C3K).
The methyl moiety of PV1322 partially occupies the GDHP as observed in prior crystallographic studies with the Chk2-specific inhibitors NSC 109555 and PV1019 [21]. The GDHP is located behind the adenine-binding region of the ATP-binding site and its accessibility by inhibitors of other kinases has been shown to be dependent on the size of the gatekeeper residue (Leu301 in Chk2) [25,32]. Large and bulky residues at this position can block access to the GDHP, whereas more compact gatekeeper residues allow bulkier substituents to be incorporated into the pocket. Furthermore, the amino acids that form the GDHPs in various kinases are not conserved and, consequently, this pocket can act as a selectivity filter for kinase inhibitors. Superposition of the coordinates of the Chk2-PV1322 complex with those of the Chk1-ABO inhibitor complex revealed that the methyl group of PV1322 and the methoxyphenol of the ABO inhibitor occupy a similar position in the GDHPs (Fig. 2b) [33]. However, the GDHPs of Chk1 and Chk2 differ. Although both kinases share a leucine residue at the gatekeeper position, in Chk2 the GDHP is lined almost entirely by hydrophobic residues whereas in Chk1 the top of the pocket is capped by a polar Asn59 residue (Leu277 in Chk2). Accordingly, we endeavored to exploit this difference between Chk2 and Chk1to design new inhibitors that would occupy the GDHP in Chk2.
3.2 Structures of Chk2 in complex with PV1352 and PV1162
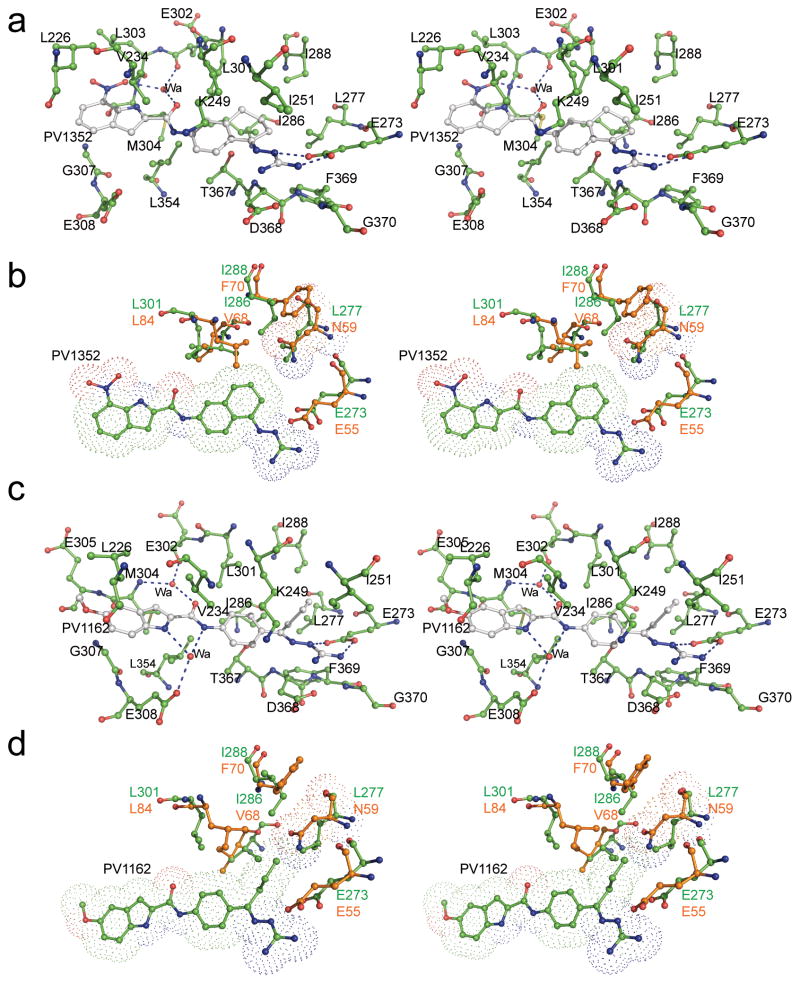
PV1019 was selected as the starting point for modification because it was more potent than PV1322 (IC50=0.16 nM, Table 1) although less selective for Chk2. Analog PV1352 (Fig. 1c) was designed by replacing the methyl group in PV1019 with a cyclohexane ring fused to the aryl ring of the phenyl bisguanidinohydrazone. PV1352 exhibited an IC50 of 0.17 nM, which is almost identical to that of PV1019, yet it was more selective than the latter compound for Chk2 over Chk1 and RSK2. Indeed, PV1352 exhibited minimal inhibition of Chk1 and its IC50 for RSK2 was much weaker than that of PV1019 (>100 μM vs. 39 μM, respectively). The 2.1 Å structure of Chk2 in complex with PV1352 showed that the core scaffold retains the same interactions that were observed in the PV1019 complex, but the newly introduced cyclohexane group now occupies the GDHP and makes favorable hydrophobic contacts with the residues that line it (Fig. 3a). The structural basis for the increased specificity of PV1352 can be inferred by superimposing the coordinates of the Chk2-PV1352 and Chk1-ABO complexes (Fig 3b), which reveals that the polar Asn59 residue of Chk1 would be in contact with the cyclohexane ring of PV1352, thereby resulting in an unfavorable polar-nonpolar interaction. Inhibitor PV1162 (Fig. 1d) was designed by replacing the 7-nitro-indole group of PV1019 with a 5-methoxy-indole and substituting the methyl group with an isobutyl moiety. This compound exhibited an IC50 of 0.29 nM for Chk2 and minimal activity against Chk1. Like PV1352, whereas PV1162 did not exhibit a substantial gain in potency, it did demonstrate a marked improvement in selectivity for Chk2 over Chk1 and RSK2. The 2.2 Å structure of Chk2 in complex with PV1162 (Fig. 3c) reveals new binding interactions around the 5-methoxy-indole moiety. Water 2042 mediates a water-bridged hydrogen-bonding network between the indole NH and the side chain carboxylate of Glu308 as well as to the nitrogen between the carbonyl and aryl ring. Several van der Waals interactions between residues Leu226, Val234, Leu303, Gly307, and the aliphatic portion of the Glu305 side chain and the 5-methoxy-indole contribute to binding interactions. The isobutyl moiety occupies but does not entirely fill the hydrophobic pocket. However, the structural overlay with the Chk1-ABO inhibitor complex suggests that, like the cyclohexane group in PV1352, unfavorable nonpolar-polar interactions between the isopropyl moiety of PV1162 and Asn59 in Chk1 along with an accompanying steric clash probably explains why PV1162 has minimal activity against Chk1. (Fig. 3d)
Table 1.
IC50 (nM) Values for inhibitors*
| Inhibitor | Chk2 | Chk1 | RSK2 |
|---|---|---|---|
| PV1322 | 12.67±11.90 | 34000 | >100000 |
| PV1019 | 0.16±0.03 | 8100 | 39000 |
| PV1352 | 0.17±0.01 | 36000** | >100000 |
| PV1162 | 0.29±0.14 | 59000** | >100000 |
Tabulated data is from head-to-head testing using a single batches of each enzyme. Chk2 values are the mean±S.D. from at least duplicate experiments. Chk1 and RSK2 determinations were single assays except as indicated of PV1352 and PV1162.
Archival assay data (mean of duplicate assays).
Fig. 3.
Stereo views of the crystal structures of (a) Chk2 (green) in complex with PV1352 (gray), (b) superimposed coordinates of Chk2-PV1352 complex (green) with Chk1-ABO complex (orange), (c) Chk2-PV1162 (gray) complex, and (d) superimposed coordinates of Chk2-PV1162 and Chk1-ABO complexes.
In summary, we have shown that despite the high degree of sequence identity in the ATP binding pockets of Chk1 and Chk2, information gleaned from co-crystal structures of enzyme-inhibitor complexes can be exploited for the rational design of highly specific inhibitors. Targeting the GDHP may be a particularly effective way to improve the specificity of Chk2 inhibitors. The structures of the inhibitor complexes described here illustrate that the key structural determinants for the binding of these compounds include a combination of water-mediated and direct hydrogen bonding to the hinge region, ionic and hydrogen bonding interactions with Glu273 that are mediated by the terminal guanidinohydrazone, and hydrophobic binding interactions with the GDHP. In addition, the structures establish a framework for further optimization of the chemical scaffold in the hinge binding region and suggest that the guanidinohydrazone moiety could be replaced with a more favorable drug-like fragment that retains interactions with Glu273. However, prior to further optimization of these inhibitors, it will be important to evaluate their pharmacological properties in cell culture [21] to determine their antiproliferative activity as single agents in cancer cells with endogenous Chk2 activation [13,21] and their synergism with DNA damaging agents in normal and p53-deficient cells [12]. The Chk2 inhibitor PV1019 meets all of these criteria [21] and we anticipate that its derivatives described here, which have been modified to target the GDHP, will perform similarly.
Supplementary Material
Highlights.
Chk2 inhibitors are designed to extend into a hydrophobic pocket in the kinase.
X-ray co-crystal structures reveal the modes of binding by the modified inhibitors.
The inhibitors occupy the hydrophobic pocket and their specificity is improved.
Acknowledgments
This project was supported in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E, the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and by the Developmental Therapeutics Program of the Division of Cancer Treatment and Diagnosis. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products or organizations imply endorsement by the US government. We thank Dr. Dominic Scudiero, Michael Selby and Julie Laudeman for conducting the kinase inhibition studies. We thank the Biophysics Resource in the Structural Biophysics Laboratory, NCI-Frederick, for use of the LC/ESMS instrument. X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) beamlines 22-ID and 22-BM, Advanced Photon Source, Argonne National Laboratory. Use of the APS was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31-109-Eng-38.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pommier Y, Weinstein JN, Aladjem MI, Kohn KW. Chk2 molecular interaction map and rationale for Chk2 inhibitors. Clin Cancer Res. 2006;12:2657–61. doi: 10.1158/1078-0432.CCR-06-0743. [DOI] [PubMed] [Google Scholar]
- 2.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–13. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 3.Stracker TH, Usui T, Petrini JH. Taking the time to make important decisions: the checkpoint effector kinases Chk1 and Chk2 and the DNA damage response. DNA Repair (Amst) 2009;8:1047–54. doi: 10.1016/j.dnarep.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn JY, Schwarz JK, Piwnica-Worms H, Canman CE. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 2000;60:5934–6. [PubMed] [Google Scholar]
- 5.Ahn JY, Li X, Davis HL, Canman CE. Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J Biol Chem. 2002;277:19389–95. doi: 10.1074/jbc.M200822200. [DOI] [PubMed] [Google Scholar]
- 6.Wu X, Chen J. Autophosphorylation of checkpoint kinase 2 at serine 516 is required for radiation-induced apoptosis. J Biol Chem. 2003;278:36163–8. doi: 10.1074/jbc.M303795200. [DOI] [PubMed] [Google Scholar]
- 7.Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–7. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J, et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol. 2004;24:708–18. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan Y, Raychaudhuri P, Costa RH. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol. 2007;27:1007–16. doi: 10.1128/MCB.01068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stolz A, et al. The CHK2-BRCA1 tumour suppressor pathway ensures chromosomal stability in human somatic cells. Nat Cell Biol. 2010;12:492–9. doi: 10.1038/ncb2051. [DOI] [PubMed] [Google Scholar]
- 11.Antoni L, Sodha N, Collins I, Garrett MD. CHK2 kinase: cancer susceptibility and cancer therapy - two sides of the same coin? Nat Rev Cancer. 2007;7:925–36. doi: 10.1038/nrc2251. [DOI] [PubMed] [Google Scholar]
- 12.Garrett MD, Collins I. Anticancer therapy with checkpoint inhibitors: what, where and when? Trends Pharmacol Sci. 2011;32:308–16. doi: 10.1016/j.tips.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 13.Zoppoli G, et al. CHEK2 genomic and proteomic analyses reveal genetic inactivation or endogenous activation across the 60 cell lines of the US National Cancer Institute. Oncogene. 2011 doi: 10.1038/onc.2011.283. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Ghamrasni S, et al. Inactivation of chk2 and mus81 leads to impaired lymphocytes development, reduced genomic instability, and suppression of cancer. PLoS Genet. 2011;7:e1001385. doi: 10.1371/journal.pgen.1001385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghosh JC, Dohi T, Raskett CM, Kowalik TF, Altieri DC. Activated checkpoint kinase 2 provides a survival signal for tumor cells. Cancer Res. 2006;66:11576–9. doi: 10.1158/0008-5472.CAN-06-3095. [DOI] [PubMed] [Google Scholar]
- 16.Gibson SL, Bindra RS, Glazer PM. CHK2-dependent phosphorylation of BRCA1 in hypoxia. Radiat Res. 2006;166:646–51. doi: 10.1667/RR0660.1. [DOI] [PubMed] [Google Scholar]
- 17.Freiberg RA, Hammond EM, Dorie MJ, Welford SM, Giaccia AJ. DNA damage during reoxygenation elicits a Chk2-dependent checkpoint response. Mol Cell Biol. 2006;26:1598–609. doi: 10.1128/MCB.26.5.1598-1609.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chabalier-Taste C, Racca C, Dozier C, Larminat F. BRCA1 is regulated by Chk2 in response to spindle damage. Biochim Biophys Acta. 2008;1783:2223–33. doi: 10.1016/j.bbamcr.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Castedo M, et al. Mitotic catastrophe constitutes a special case of apoptosis whose suppression entails aneuploidy. Oncogene. 2004;23:4362–70. doi: 10.1038/sj.onc.1207572. [DOI] [PubMed] [Google Scholar]
- 20.Vakifahmetoglu H, Olsson M, Tamm C, Heidari N, Orrenius S, Zhivotovsky B. DNA damage induces two distinct modes of cell death in ovarian carcinomas. Cell Death Differ. 2008;15:555–66. doi: 10.1038/sj.cdd.4402286. [DOI] [PubMed] [Google Scholar]
- 21.Jobson AG, et al. Cellular inhibition of checkpoint kinase 2 (Chk2) and potentiation of camptothecins and radiation by the novel Chk2 inhibitor PV1019 [7-nitro-1H-indole-2-carboxylic acid {4-[1-(guanidinohydrazone)-ethyl]-phenyl}-amide] J Pharmacol Exp Ther. 2009;331:816–26. doi: 10.1124/jpet.109.154997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takai H, et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. Embo J. 2002;21:5195–205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arienti KL, et al. Checkpoint kinase inhibitors: SAR and radioprotective properties of a series of 2-arylbenzimidazoles. J Med Chem. 2005;48:1873–85. doi: 10.1021/jm0495935. [DOI] [PubMed] [Google Scholar]
- 24.Carlessi L, Buscemi G, Larson G, Hong Z, Wu JZ, Delia D. Biochemical and cellular characterization of VRX0466617, a novel and selective inhibitor for the checkpoint kinase Chk2. Mol Cancer Ther. 2007;6:935–44. doi: 10.1158/1535-7163.MCT-06-0567. [DOI] [PubMed] [Google Scholar]
- 25.Toledo LM, Lydon NB, Elbaum D. The structure-based design of ATP-site directed protein kinase inhibitors. Curr Med Chem. 1999;6:775–805. [PubMed] [Google Scholar]
- 26.Scapin G. Structural biology in drug design: selective protein kinase inhibitors. Drug Discov Today. 2002;7:601–11. doi: 10.1016/s1359-6446(02)02290-0. [DOI] [PubMed] [Google Scholar]
- 27.Lountos GT, Tropea JE, Zhang D, Jobson AG, Pommier Y, Shoemaker RH, Waugh DS. Crystal structure of checkpoint kinase 2 in complex with NSC 109555, a potent and selective inhibitor. Protein Sci. 2009;18:92–100. doi: 10.1002/pro.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–66. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 29.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–5. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 30.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–95. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 31.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–83. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang D, Zhou T, Lafleur K, Nevado C, Caflisch A. Kinase selectivity potential for inhibitors targeting the ATP binding site: a network analysis. Bioinformatics. 2010;26:198–204. doi: 10.1093/bioinformatics/btp650. [DOI] [PubMed] [Google Scholar]
- 33.Foloppe N, Fisher LM, Francis G, Howes R, Kierstan P, Potter A. Identification of a buried pocket for potent and selective inhibition of Chk1: prediction and verification. Bioorg Med Chem. 2006;14:1792–804. doi: 10.1016/j.bmc.2005.10.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.