Abstract
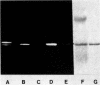
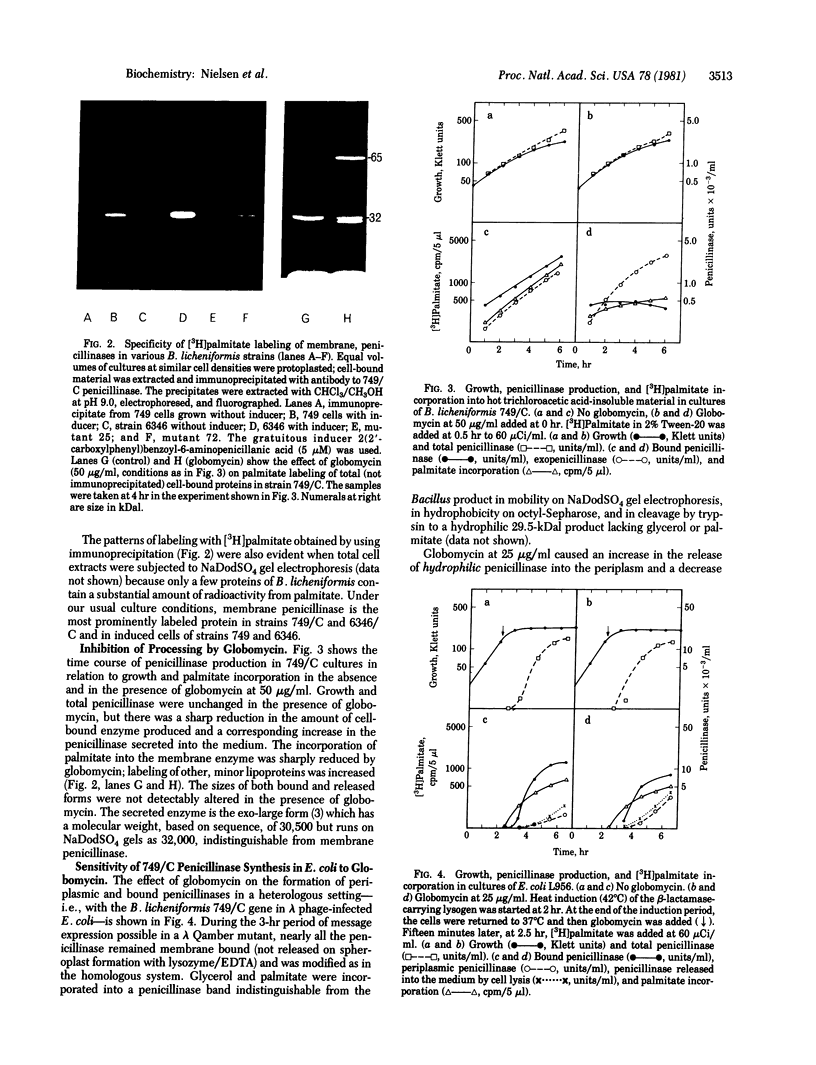
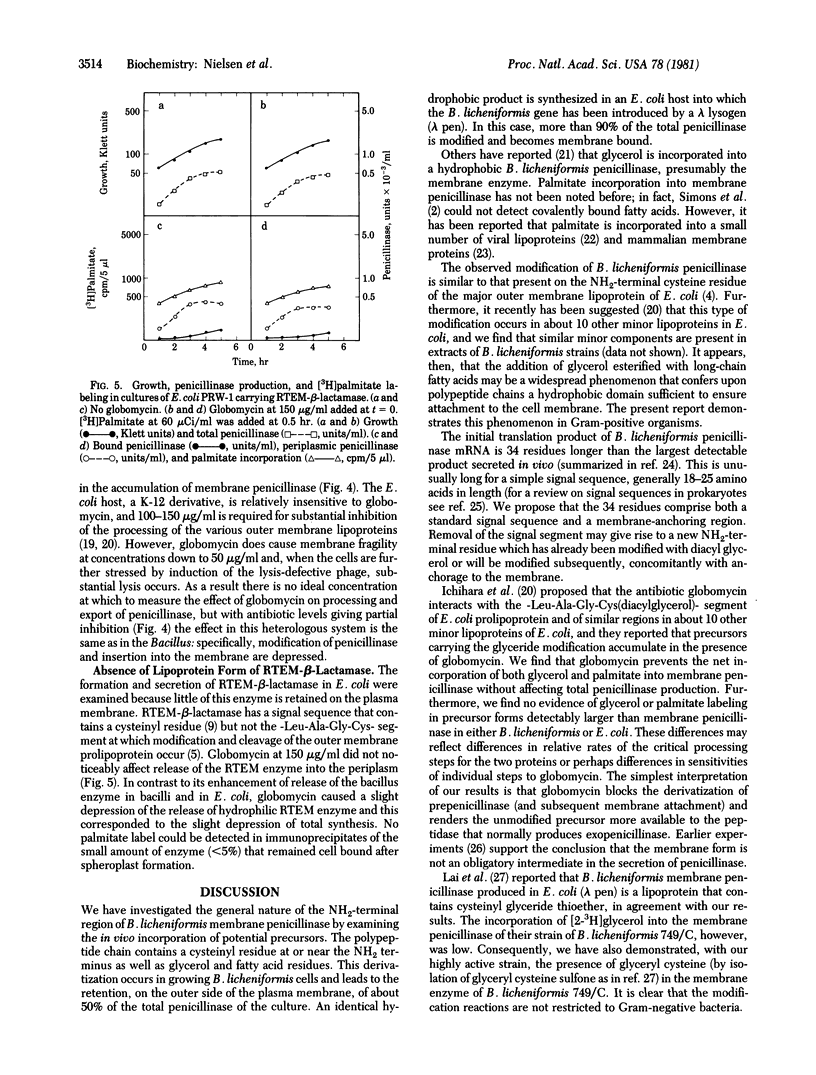
Membrane penicillinase (penicillin amido-beta-lactamhydrolase, EC 3.5.2.6) from Bacillus licheniformis bears a striking resemblance to the major outer membrane lipoprotein of Escherichia coli. It can be specifically labeled in vivo with [3H]glycerol, [35S]cysteine, or [3H]palmitate but not by [32P]orthophosphate. The labeled residues are located at or near the NH2 terminus of the membrane penicillinase because they can be completely removed by trypsin which cleaves a hydrophobic peptide(s) from the NH2 terminus, thereby rendering the enzyme hydrophilic. The membrane penicillinase produced by the 749/C gene carried in E. coli on phage lambda is similar to the enzyme formed in strain 749/C itself. The peptide antibiotic globomycin, which prevents processing of the E. coli prolipoprotein, severely inhibited the attachment of [3H]palmitate or [3H]glycerol to the 749/C enzyme (either in B. licheniformis 749/C or in E. coli), blocked the accumulation of penicillinase in the plasma membrane, and enhanced the formation of exoenzyme. Under the same conditions, globomycin does not prevent the attachment of palmitate or glycerol to the E. coli prolipoprotein but inhibits processing of the modified precursor to the mature lipoprotein. These results are in contrast with the lack of effect of globomycin on the RTEM-beta-lactamase of E. coli which has no detectable hydrophobic membrane form and was not labeled with palmitate or glycerol.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P., Meadway R. J. Chemical structure of bacterial penicillinases. Nature. 1969 Apr 5;222(5188):24–26. doi: 10.1038/222024a0. [DOI] [PubMed] [Google Scholar]
- BLIGH E. G., DYER W. J. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959 Aug;37(8):911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Birdsell D. C., Cota-Robles E. H. Production and ultrastructure of lysozyme and ethylenediaminetetraacetate-lysozyme spheroplasts of Escherichia coli. J Bacteriol. 1967 Jan;93(1):427–437. doi: 10.1128/jb.93.1.427-437.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brammar W. J., Muir S., McMorris A. Molecular cloning of the gene for the beta-lactamase of Bacillus licheniformis and its expression in Escherichia coli. Mol Gen Genet. 1980 Apr;178(1):217–224. doi: 10.1007/BF00267232. [DOI] [PubMed] [Google Scholar]
- Chamberlain J. P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979 Sep 15;98(1):132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Crane L. J., Bettinger G. E., Lampen J. O. Affinity chromatography purification of penicillinase of Bacillus licheniformis 749-C and its use to measure tuurnover of the cell bound enzyme. Biochem Biophys Res Commun. 1973 Jan 23;50(2):220–227. doi: 10.1016/0006-291x(73)90829-2. [DOI] [PubMed] [Google Scholar]
- Emr S. D., Hall M. N., Silhavy T. J. A mechanism of protein localization: the signal hypothesis and bacteria. J Cell Biol. 1980 Sep;86(3):701–711. doi: 10.1083/jcb.86.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halegoua S., Hirashima A., Sekizawa J., Inouye M. Protein synthesis in toluene-treated Escherichia coli. Exclusive synthesis of membrane proteins. Eur J Biochem. 1976 Oct 1;69(1):163–167. doi: 10.1111/j.1432-1033.1976.tb10869.x. [DOI] [PubMed] [Google Scholar]
- Halegoua S., Inouye M. Translocation and assembly of outer membrance proteins of Escherichia coli. Selective accumulation of precursors and novel assembly intermediates caused by phenethyl alcohol. J Mol Biol. 1979 May 5;130(1):39–61. doi: 10.1016/0022-2836(79)90551-5. [DOI] [PubMed] [Google Scholar]
- Hantke K., Braun V. Covalent binding of lipid to protein. Diglyceride and amide-linked fatty acid at the N-terminal end of the murein-lipoprotein of the Escherichia coli outer membrane. Eur J Biochem. 1973 Apr;34(2):284–296. doi: 10.1111/j.1432-1033.1973.tb02757.x. [DOI] [PubMed] [Google Scholar]
- Inukai M., Takeuchi M., Shimizu K., Arai M. Mechanism of action of globomycin. J Antibiot (Tokyo) 1978 Nov;31(11):1203–1205. doi: 10.7164/antibiotics.31.1203. [DOI] [PubMed] [Google Scholar]
- Izui K., Nielsen J. B., Caulfield M. P., Lampen J. O. Large exopenicillinase, initial extracellular form detected in cultures of Bacillus licheniformis. Biochemistry. 1980 Apr 29;19(9):1882–1886. doi: 10.1021/bi00550a023. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Lai J. S., Sarvas M., Brammar W. J., Neugebauer K., Wu H. C. Bacillus licheniformis penicillinase synthesized in Escherichia coli contains covalently linked fatty acid and glyceride. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3506–3510. doi: 10.1073/pnas.78.6.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampen J. O., Nielsen J. B., Izui K., Caulfield M. P. Bacillus licheniformis beta-lactamases: multiple forms and their roles. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):345–348. doi: 10.1098/rstb.1980.0051. [DOI] [PubMed] [Google Scholar]
- POLLOCK M. R. PURIFICATION AND PROPERTIES OF PENICILLINASES FROM TWO STRAINS OF BACILLUS LICHENIFORMIS: A CHEMICAL, PHYSICOCHEMICAL AND PHYSIOLOGICAL COMPARISON. Biochem J. 1965 Mar;94:666–675. doi: 10.1042/bj0940666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POLLOCK M. R. STIMULATING AND INHIBITING ANTIBODIES FOR BACTERIAL PENICILLINASE. Immunology. 1964 Nov;7:707–723. [PMC free article] [PubMed] [Google Scholar]
- Sargent M. G. Rapid fixed-time assay for penicillinase. J Bacteriol. 1968 Apr;95(4):1493–1494. doi: 10.1128/jb.95.4.1493-1494.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger M. J., Magee A. I., Schmidt M. F. Fatty acid acylation of proteins in cultured cells. J Biol Chem. 1980 Nov 10;255(21):10021–10024. [PubMed] [Google Scholar]
- Schmidt M. F., Bracha M., Schlesinger M. J. Evidence for covalent attachment of fatty acids to Sindbis virus glycoproteins. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1687–1691. doi: 10.1073/pnas.76.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherratt D. J., Collins J. F. Analysis by transformation of the penicillinase system in Bacillus licheniformis. J Gen Microbiol. 1973 May;76(1):217–230. doi: 10.1099/00221287-76-1-217. [DOI] [PubMed] [Google Scholar]
- Simons K., Sarvas M., Garoff H., Helenius A. Membrane-bound and secreted forms of penicillinase from Bacillus licheniformis. J Mol Biol. 1978 Dec 25;126(4):673–690. doi: 10.1016/0022-2836(78)90015-3. [DOI] [PubMed] [Google Scholar]
- Smith W. P., Tai P. C., Davis B. D. Bacillus licheniformis penicillinase: cleavages and attachment of lipid during cotranslational secretion. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3501–3505. doi: 10.1073/pnas.78.6.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S., Lampen J. O. The hydrophobic membrane penicillinase of Bacillus licheniformis 749/C. Characterization of the hydrophilic enzyme and phospholipopeptide produced by trypsin cleavage. J Biol Chem. 1976 Jul 10;251(13):4102–4110. [PubMed] [Google Scholar]