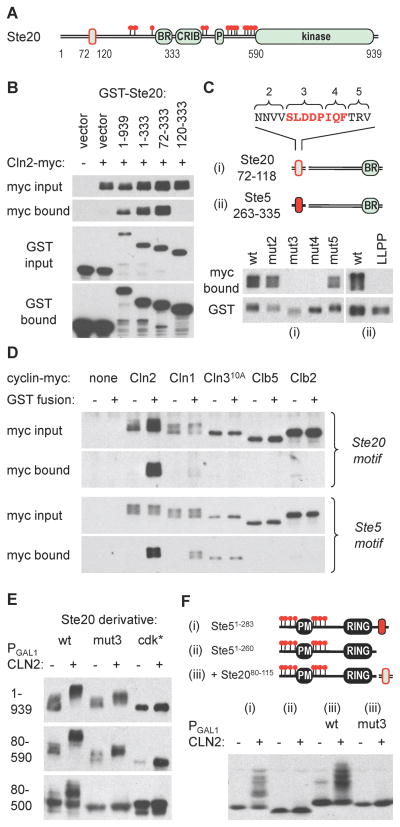
Figure 3. Cyclin-specific binding by docking motifs in Ste20 and Ste5.
(A) The diagram indicates endpoints used for mapping the Cln2-binding region in Ste20, which is outlined in red
(B) Cells co-expressing GST fusions to Ste20 fragments and Cln2-myc13 were lysed, and complexes were recovered using glutathione sepharose. Input (5%) and bound proteins were analyzed by anti-myc and anti-GST blots
(C) Starting with a Ste2072–333 fragment, alanine substitutions were made at blocks of residues in the 72–118 region (see Figure S2C; numbering starts at 2 because additional flanking mutations were used in other assays). Cln2 binding was tested as in panel B. Separately, the required Ste20 region was replaced by a Ste5263–335 fragment (ii), in both wt and LLPP mutant forms, to test the ability of this Ste5 sequence to mediate Cln2 binding
(D) Ste20 and Ste5 docking sites show cyclin-specific binding. GST alone (−) or GST fusions (+) were used to co-precipitate myc13-tagged cyclins (expressed from the CYC1 promoter) in yeast lysates. The GST fusions were to Ste201–333 (Ste20 motif) or to the Ste5263–335-Ste20120–333 chimera used in panel Cii (Ste5 motif). Cln310A showed varying levels of non-specific precipitation but no reproducible binding to either GST fusion
(E) Cln2-induced phosphorylation was assayed for V5-tagged forms of full-length Ste20 (1–939) or N-terminal fragments (80–590, 80–500), with or without mutations in the docking site (mut3) or the 13 confirmed Cdk sites (cdk*)
(F) The Cln2 docking site from Ste20 can drive phosphorylation of a heterologous substrate. Phosphorylation was analyzed using Ste51–283 (i), Ste51–260 (ii), and wt or mut3 versions of the Ste20 docking site (residues 80–115) fused to Ste51–260 (iii).