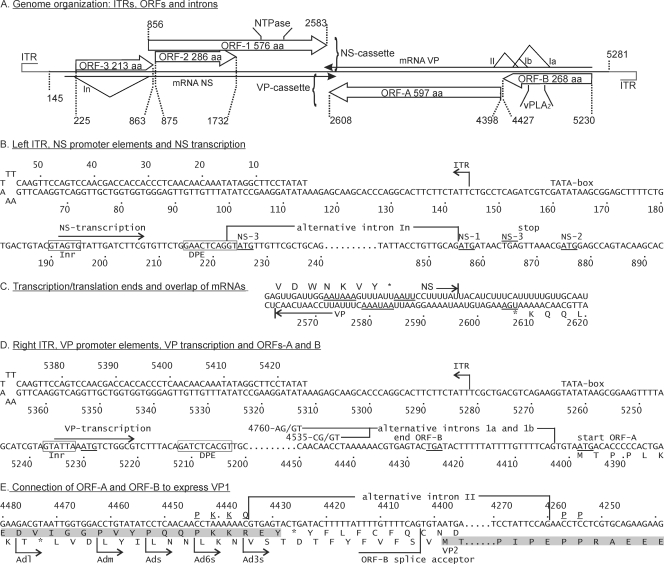
Fig. 1.
Genome organization of AdDNV. (A) Overview of genome organization, positions, and sizes of ITRs, ORFs, and introns. The In intron in the NS mRNA, between nt 223 and nt 855, occurs in about half of the NS transcripts. Ia (nt 4403 to 4758), Ib (nt 4403 to 4533), and II (nt 4260 to 4434) are introns that occur in alternative VP transcripts. vPLA2 indicates the position of the viral phospholipase A2 motif. (B) Left ITR and regulation of production of NS transcripts. NS transcripts start at 192-A and yield NS3 from 225-A. However, a fraction of these transcripts are spliced just upstream of this start codon (intron In), leading to translation of NS1 from 856-A (AUG with a poor initiation environment) and, through leaky scanning, of NS2 from 875-A. Inr and DPE are promoter elements. (C) Like the case for all members of the Densovirus genus, the 3′ ends of AdDNV NS and VP transcripts overlap in the middle of the genome. The stop codons and AATAAA motifs are underlined. (D) Right ITR, VP transcription sites, and splicing in ORF-B on the complementary strand. Transcription starts at nt 5235, and VP1 initiation is at nt 5230. The short 5′-UTR predicts an inefficient initiation (leaky scanning) and could be responsible for the production of a nested set of N-terminally extended viral proteins. However, removal of either of the two alternative introns in ORF-B (Ia or Ib) did not connect the exons in ORF-B and ORF-A in frame, so only nonstructural proteins could be produced from nt 5230 and VP2 could be produced directly from the first AUG in ORF-A when this splicing occurred. (E) An alternative intron II, which is mutually exclusive with introns Ia and Ib because the ORF-B splice acceptor is removed, connects ORF-B and ORF-A (both shaded) in frame so that VP1 can be produced from nt 5230. The VP1 sequence around the splicing site is underlined and shown above the nt sequence.