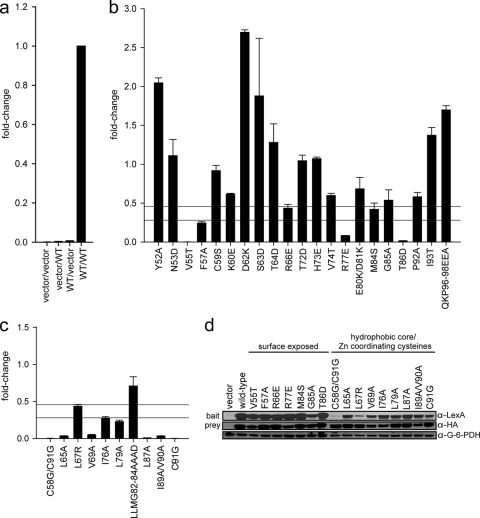
Fig. 2.
E7 dimerization in vivo. (a) Controls for the yeast two-hybrid assay. L-40 yeast cells were cotransformed with a LexA DNA binding domain (bait) and a B42 activation domain (prey). The combination of vectors (bait/prey) used is indicated below each data bar, with “vector” representing an empty vector and “WT” indicating a vector expressing a wild-type HPV16 E7 fusion. Indicated constructs were expressed in L-40 yeast cells containing an integrated LexA-responsive β-galactosidase reporter. Cell extracts were prepared and assayed for β-galactosidase activity. (b) Dimerization of mutants with mutations targeting predicted surface-exposed residues as determined by the yeast two-hybrid assay. HPV16 E7 mutants were expressed as fusions to the LexA DNA binding domain and the B42 activation domain in L-40 yeast cells, and β-galactosidase activity was assessed. The upper horizontal line is set at 45% of wild-type activity, corresponding to a predicted 5-fold increase in Kd; the lower horizontal line corresponds to a predicted 10-fold increase in Kd and is set at 28% of wild-type activity. We considered any mutation with <45% of wild-type activity to be impaired for dimerization. The same criterion also applies to panel c. (c) Dimerization of mutants with mutations targeting predicted hydrophobic core residues and zinc-coordinating cysteines as assessed by the yeast two-hybrid assay. (d) Expression levels of LexA and B42 fusion proteins of dimerization-impaired E7 mutants. Yeast extracts cotransformed with either wild-type E7 or the E7 mutant as both LexA (bait) and B42 (prey) fusions were tested for protein expression by Western blotting using anti-LexA and anti-HA antibodies. An anti-G6PD blot served as a loading control.