Abstract
Viruses can modify conditions inside cells to make them more favorable for replication and progeny virus production. One way of doing this is through manipulation of the cell cycle, a process that describes the ordered growth and division of cells. Analysis of model cell lines, such as A549 cells and primary airway epithelial cells, infected with human respiratory syncytial virus (HRSV) has shown alteration of the cell cycle during infection, although the signaling events were not clearly understood. In this study, targeted transcriptomic analysis of HRSV-infected primary airway epithelial cells revealed alterations in the abundances of many mRNAs encoding cell cycle-regulatory molecules, including decreases in the D-type cyclins and corresponding cyclin-dependent kinases (CDK4 and CDK6 [CDK4/6]). These alterations were reflected in changes in protein abundance and/or relocalization in HRSV-infected cells; taken together, they were predicted to result in G0/G1 phase arrest. In contrast, there was no change in the abundances of D-type cyclins in A549 cells infected with HRSV. However, the abundance of the G1/S phase progression inhibitor p21WAF1/CIP1 was increased over that in mock-treated cells, and this, again, was predicted to result in G0/G1 phase arrest. The G0/G1 phase arrest in both HRSV-infected primary cells and A549 cells was confirmed using dual-label flow cytometry that accurately measured the different stages of the cell cycle. Comparison of progeny virus production in primary and A549 cells enriched in G0/G1 using a specific CDK4/6 kinase inhibitor with asynchronously replicating cells indicated that this phase of the cell cycle was more efficient for virus production.
INTRODUCTION
Interaction with the cell cycle is one way in which viruses can create favorable conditions for enhancing virus replication and progeny virus production (5). The cell cycle is separated into several distinct phases that describe the ordered growth and division of a cell and reflect the different metabolic processes occurring at particular time points. In the first gap phase (G1), translation is maximal in the cell, as reflected in the increased number of nucleoli producing ribosomal subunits. This is followed by the synthesis (S) phase, where DNA is replicated, with the content increasing from 2N to 4N. Cells then enter the G2 phase, another round of protein synthesis, prior to entry into the mitosis (M) phase, in which nuclear division is followed by cytokinesis. Cells can then exit the cell cycle and enter the G0 phase. Progression through these phases is controlled by positive regulators, including the cyclins and their partner molecules, cyclin-dependent kinases (CDKs). These include the D-type cyclins (e.g., found in association with CDK4/CDK6 [CDK4/6]), which enable progression through G1; cyclins A and E (both associated with CDK2), which enable entry into and progression through S phase; and cyclin B (associated with CDK1), which enables entry into and progression through M phase. Several other molecules are critically important for cell cycle progression, including the tumor suppressor protein p53 and retinoblastoma (Rb) protein. At each stage of the cell cycle, negative regulators can act to delay or inhibit progression by controlling the protein activities, abundances, localization, and mRNA levels of different positive regulators. One example of this type of molecule is p21WAF1/CIP1, which directly inhibits the activities of cyclin D/CDK4 and cyclin E/CDK2 complexes.
Viruses and their proteins can interact with the molecules involved in cell cycle regulation to either delay, arrest, or advance this process so as to alter the host cell and/or increase progeny virus production. For example, DNA viruses, including herpesviruses, can encode cell cycle-regulatory proteins (2, 24), including homologs to cyclins (31, 39), that result in cell cycle arrest in virus-infected cells (24). Retroviruses, such as HIV-1, can induce G2/M phase arrest through manipulation of cyclin B (among other molecules) to promote virus infection (1, 13, 18, 20, 35). Viruses with RNA genomes can also interact with the cell cycle, including viruses whose site of replication is the cytoplasm. For example, in measles virus-infected cells, a block in the G0 phase (and hence in proliferation) is thought to lead to immunosuppression (30, 36) through the degradation of proteins associated with the G1 phase transition (10), possibly induced by the measles virus nucleoprotein (21). Avian coronavirus-infected cells are arrested in the G2/M phase (8, 22), where nucleolar trafficking of the virally encoded nucleocapsid protein is greater (3) and progeny virus production is more efficient (8, 22). Infection of cells with murine coronavirus results in G0/G1 phase arrest (4). Influenza virus, whose site of RNA genome replication and transcription is the nucleus, has also been shown to induce cell cycle arrest and changes in the abundances of cell cycle-regulatory proteins (19, 33).
Human respiratory syncytial virus (HRSV), a negative-sense RNA virus belonging to the order Mononegavirales, has been reported to cause G2/M phase arrest in primary human bronchial epithelial (PHBE) cells (12), a target of infection in vivo, and G0/G1 phase arrest in the continuous A549 (12) and HEp-2 (26) cell lines. A549 cells are derived from a human male and are adenocarcinomic human alveolar basal epithelial cells, with a modal chromosome number of 66. Thus, the differences in cell cycle phenotypes between primary and continuous cells could be due to genotypic and cell type differences (12) and emphasize the relevance of primary cells for these studies.
HRSV has been proposed to induce cell cycle arrest by activating transforming growth factor β (12) and by decreasing the level of p53 to prolong the survival of airway epithelial cells (14). Expression of the virus-encoded fusion (F) protein has been proposed to induce p53-dependent apoptosis in A549 cells but not in HEp-2 cells (which express lower levels of p53) (9). In quantitative proteomic analyses of A549 cells infected with either HRSV subgroup A or subgroup B, the abundances of several different cell cycle-regulatory molecules have been shown to change at 24 h postinfection (27, 28), consistent with cell cycle dysfunction (12). However, the molecular signaling events associated with HRSV and the cell cycle in both primary cells and continuous cell culture have not been resolved.
In the current study, the regulation of molecules involved in cell cycle control at both the mRNA and the protein level in HRSV-infected cells was investigated principally in primary human airway (bronchial/tracheal) epithelial cells (HBEpC/HTEpC; referred to below as HBEpC cells) but also in continuous cell culture (A549 cells). Although there were distinct differences between the cell types in the regulation of cell cycle-regulatory molecules due to HRSV infection, all were predicted to arrest the cell cycle in the G0/G1 phase, which was confirmed experimentally. Cell cycle enrichment studies also demonstrated that this stage of the cell cycle resulted in increased progeny virus production over that for HRSV grown in asynchronously replicating cells.
MATERIALS AND METHODS
Cells and virus.
Primary human bronchial epithelial cells (HBEpC cells) and A549 cells were obtained from Health Protection Agency Culture Collections (HPACC). HBEpC cells were grown in serum-free bronchial epithelial basal medium with growth supplement. Fresh medium was supplied every 2 days, and cells were used within 16 population doublings (approximately 4 passages). A549 cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum (FBS). HRSV strain A2 was kindly provided by Patricia Cane (HPA) and was passaged in HEp-2 cells. An infectious viral stock and a mock viral stock (medium with cell lysate) were prepared in parallel, snap-frozen, and stored at −80°C. Viral titers were determined in HEp-2 cells by a modified methylcellulose plaque assay using a goat anti-HRSV antibody (Abcam). For all experiments, unless otherwise specified, cells were infected with HRSV at a multiplicity of infection (MOI) of 1.
Transcriptomic analysis.
HBEpC cells were either mock infected or infected with HRSV for 24 h. The total RNAs of mock-infected and infected cells were extracted using the RNeasy Mini kit (Qiagen). Genomic DNA contamination was eliminated using the RNase-free DNase set (Qiagen). cDNA was synthesized from 1 μg of total RNA using the ThermoScript reverse transcription-PCR (RT-PCR) system (Invitrogen). After successful RT was confirmed by PCRs with primers of human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or the HRSV G gene, quantitative real-time PCR was performed using the Human Cell Cycle RT2 Profiler PCR array (PAHS-020A; SABiosciences) on an ABI 7900HT real-time PCR system (Applied Biosystems) according to the manufacturer's instructions. Data were processed using the Excel-based PCR Array Data Analysis software (SABiosciences).
Bioinformatic analysis of cell cycle deregulation.
Data were analyzed by the use of Ingenuity Pathway Analysis software (Ingenuity Systems). Networks were generated using data sets containing gene identifiers and corresponding expression values, which were uploaded into the application. Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base. An enrichment level of 2.0 or greater was arbitrarily set to identify the genes whose change in abundance may have functional relevance. These genes, called focus genes, were overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base. Networks of these focus genes were then algorithmically generated based on their connectivity. Graphical representations of the molecular relationships between genes/gene products were generated. Genes or gene products are represented as nodes, and the biological relationship between two nodes is represented as an edge (line). All edges are supported by at least one reference from the literature or from canonical information stored in the Ingenuity Pathways Knowledge Base. Human, mouse, and rat orthologs of a gene are stored as separate objects in the Ingenuity Pathways Knowledge Base but are represented as a single node in the network. The intensity of the node color indicates the relative change in expression level. Nodes are displayed using various shapes that represent the functional class of the gene product. The P value associated with functions and pathways in Ingenuity Pathway Analysis is a measure of the likelihood that the association between a set of focus molecules in the experiment and a given function/pathway is due to random chance. The smaller the P value, the less likely that the association is random, and the more significant the association. The P values are calculated using the right-tailed Fisher exact test, and in general, P values less than 0.05 indicate a significant, nonrandom association.
Flow cytometry analysis of the cell cycle.
At 12 h and 24 h postinfection, 10 μM 5-ethynyl-2′-deoxyuridine (EdU) was added to the culture medium and was incubated for 30 min in order to incorporate it into the synthesizing DNA. Cells were then harvested with immediate fixation and were dual labeled using the Click-iT EdU flow cytometry assay kit (Invitrogen) by following the manufacturer's instructions. The EdU label was detected with Alexa Fluor 647 based on a click reaction under mild conditions to accurately determine the proportion of cells in S phase. DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI) to measure cells in the G0/G1 (2N) and G2/M (4N) phases. Additionally, HRSV was labeled with a fluorescein isothiocyanate (FITC)-conjugated goat anti-HRSV antibody (Abcam) to monitor the infection. Cell cycle distribution was analyzed by the LSRFortessa cell analyzer (BD Biosciences).
Immunofluorescence microscopy.
HBEpC cells were grown on coverslips and were either mock infected or HRSV infected. At 12 h and 24 h postinfection, the coverslips were washed with phosphate-buffered saline (PBS)–0.5% Tween 20 (PBST) and were fixed in 4% formaldehyde. Cells were permeabilized with 0.1% Triton X-100 in PBS and were blocked with 5% FBS in PBST for 1 h. Primary antibodies against cyclin D1, cyclin D2, and HRSV were diluted 1:100 in PBST containing 2% FBS for 1 h and were detected with an Alexa Fluor 546-conjugated donkey anti-rabbit secondary antibody from Invitrogen or an FITC-conjugated donkey anti-goat secondary antibody (ab6881) from Abcam, both diluted 1:200 in PBST containing 2% FBS for 1 h. Coverslips were washed with PBST and were mounted on glass microscope slides using ProLong Gold mounting medium with DAPI (Invitrogen). Confocal images were captured on an LSM 510 META inverted confocal microscope (Carl Zeiss Ltd.).
Inhibition studies.
The proteasome inhibitor MG-132 (Calbiochem), the CRM-1-dependent nuclear export inhibitor leptomycin B (LMB; Calbiochem), and the glycogen synthase kinase 3β (GSK3β) activity inhibitor lithium chloride (LiCl; Sigma) were used to study the degradation pathway of D-type cyclins. HBEpC cells seeded in 6-well plates with 70% confluence were either mock treated or HRSV infected. At 20 h postinfection, cells were treated with either 5 μM MG-132, 2.5 ng/ml LMB, or 5 mM LiCl until 24 h postinfection. Cells were lysed with radioimmunoprecipitation assay (RIPA) buffer, and Western blot analysis was performed using an antibody against cyclin D1, cyclin D2, GAPDH, or HRSV where indicated. To enrich HBEpC and A549 cells in the G1 phase, CDK4/6 inhibitor IV (catalog no. 219492; Calbiochem), which effectively inhibits the phosphorylation of the CDK4 substrate pRb to prevent G1/S transition, was used. HBEpC and A549 cells were either mock treated or HRSV infected and were treated with 10 μM CDK4/6 inhibitor IV for 1 h before infection and throughout 24 h postinfection.
Immunoblot analysis.
Whole-cell lysates were prepared using RIPA buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], Roche protease inhibitor cocktail). The protein concentration was determined using the bicinchoninic acid (BCA) assay (Pierce). Five micrograms of protein extract was separated by 10% SDS-polyacrylamide gel electrophoresis (PAGE) gels and was transferred to polyvinylidene difluoride (PVDF) membranes (Millipore) by semidry electroblotting. The membranes were blocked in Tris-buffered saline (TBS)–0.1% Tween containing 5% dry skim milk (Invitrogen) for 1 h and were probed with primary antibodies against cyclin A (sc-751), cyclin B1 (sc-7393), cyclin D1 (sc-718), cyclin D2 (sc-593), cyclin E (sc-481), p21waf1/cip1 (sc-397), or CDK4 (sc-601) (all from Santa Cruz) or against CDK1 (ab18), HRSV (ab20745), or GAPDH (ab8245) (all from Abcam). Bound primary antibodies were detected with appropriate horseradish peroxidase (HRP)-conjugated goat anti-mouse/anti-rabbit (Invitrogen) or rabbit anti-goat (Abcam) secondary antibodies, followed by enhanced chemiluminescence.
RESULTS
HRSV infection resulted in decreased abundances of cyclin D1 and D2 proteins in HBEpC cells but not in A549 cells.
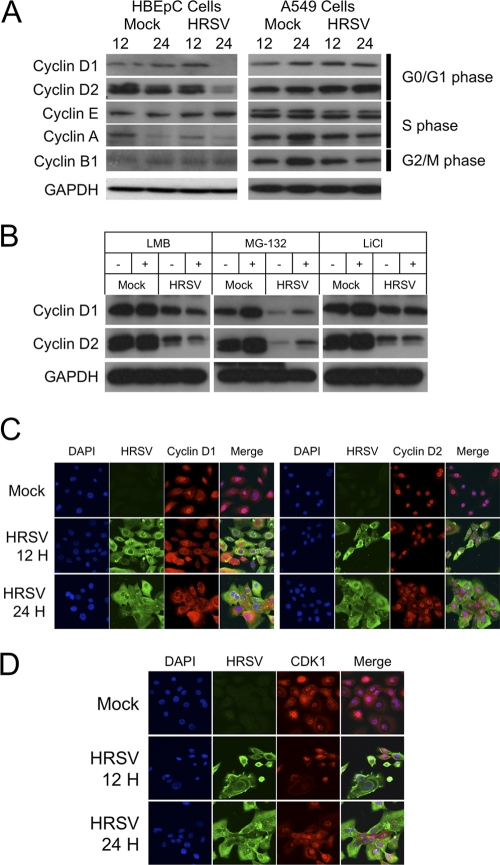
Alterations of the cell cycle in HRSV-infected cells were predicted to result in possible changes in the abundances of the major cyclins associated with cell cycle progression. Changes in the abundances of cyclin proteins would be predicted to reflect the stage of cell cycle arrest/delay in HRSV-infected cells. This was tested by immunoblotting for cyclin D1 and D2 proteins (G1 phase), cyclin A and E proteins (S phase), and cyclin B1 proteins (G2/M phase) at 12 and 24 h after infection with HRSV and comparing the results to those for mock-treated cells at 12 and 24 h in order to control for possible cell cycle effects due to density and contact inhibition (Fig. 1A). The data indicated that the abundances of both cyclin D1 and D2 proteins were decreased in HRSV-infected HBEpC cells but not in HRSV-infected A549 cells. The abundance of cyclin A protein was decreased in both mock-treated and HRSV-infected HBEpC cells at 24 h compared to 12 h. For both cell types, there was no difference in the abundance of cyclin E protein between mock-treated and HRSV-infected cells. Cyclin B1 protein showed no change in abundance in HBEpC cells, but its abundance in HRSV-infected A549 cells was lower than that in mock-treated cells at 24 h postinfection.
Fig. 1.
(A) Immunoblot analysis of the abundances of major cyclins involved in cell cycle progression in mock-treated and HRSV-infected primary HBEpC or A549 cells at 12 and 24 h postinfection. GAPDH was used to show equal protein loading, and the stage of the cell cycle associated with each cyclin is given on the right. (B) Immunoblot analysis of the abundances of cyclins D1 and D2 in HBEpC cells that were either mock treated or infected with HRSV and were grown in the absence (−) or presence (+) of an inhibitor (LMB, MG-132, or LiCl) that can act on D-type cyclin degradation. (C) Analysis by indirect immunofluorescence confocal microscopy of the localizations of cyclins D1 and D2 in mock-treated and HRSV-infected primary HBEpC cells at 12 and 24 h postinfection. The nucleus is stained blue with DAPI; viral proteins are shown in green; D-type cyclins are shown in red; and a merge image is presented. (D) Analysis by indirect immunofluorescence confocal microscopy of the localization of CDK1 in mock-treated and HRSV-infected primary HBEpC cells at 12 and 24 h postinfection. The nucleus is stained blue with DAPI; viral proteins are shown in green; CDK1 is shown in red; and a merge image is presented.
Degradation of cyclin D1 and D2 proteins in HBEpC cells.
In growing and dividing cells, cyclin D1 protein can be phosphorylated on Thr286 by glycogen synthase kinase 3β (GSK3β), promoting nuclear export and subsequent degradation in the cytoplasm (6, 15). However, cyclin D1 protein can also be exported for degradation independently of GSK3β via unknown mechanisms (7). To investigate these possibilities, HBEpC cells were grown in the presence of various inhibitors–MG-132, LiCl, or LMB (to inhibit the export and degradation of cyclin D protein)–at concentrations that have a functional effect (17) and were either mock treated or infected with HRSV. The amounts of cyclin D1 and D2 proteins were then determined by immunoblotting in relation to the amount of GAPDH at 24 h postinfection (Fig. 1B). To test whether the degradation of cyclin D1 and D2 proteins in HRSV-infected HBEpC cells was mediated via the cellular proteasomal pathway, cells were either mock treated or infected with HRSV and were then treated with the proteasomal inhibitor MG-132. The data indicated that the abundances of cyclin D1 and D2 proteins were greater in HRSV-infected HBEpC cells treated with MG-132 than in HRSV-infected cells grown in the absence of MG-132. However, these amounts were still less than the abundances of cyclin D1 and D2 proteins in mock-treated HBEpC cells either in the absence or in the presence of the inhibitor (the latter resulted in larger amounts of cyclin D1 and D2 proteins). The inhibitor LiCl was used to prevent GSK3β activity, and LMB was used to inhibit CRM-1-dependent nuclear export. Neither of these inhibitors affected the decrease in the abundances of cyclin D1 and D2 proteins in HRSV-infected HBEpC cells from those in mock-treated cells. Taken together, these findings indicated that cyclin D1 and D2 proteins were degraded by the proteasome but that this degradation also involved GSK3β-independent routes.
The subcellular localizations of cyclin D1 and D2 proteins were altered in HRSV-infected HBEpC cells.
The reductions in the abundances of D-type cyclin proteins in virus-infected cells can also be related to changes in subcellular localization (17). To investigate this in the context of HRSV-infected cells, the subcellular localizations of cyclin D1 and D2 proteins were determined using indirect immunofluorescence confocal microscopy of HRSV-infected HBEpC cells at 12 and 24 h postinfection and were compared to those for mock-treated cells (Fig. 1C). The data indicated that in mock-treated cells, cyclin D1 protein was localized predominately to the nucleus and cyclin D2 protein was exclusively nuclear. There was no obvious change in the subcellular localization of CDK1 protein (Fig. 1D). However, in HRSV-infected cells, cyclin D1 and D2 proteins were also observed in the cytoplasm, indicating that virus infection resulted in changes in the localizations of these proteins.
Transcriptomic analysis of mRNA levels encoding cell cycle-regulatory molecules in HRSV-infected HBEpC cells revealed changes associated with the G1/S phase transition.
To investigate potential changes in the levels of mRNAs encoding cell cycle-regulatory proteins, the RT2 Profiler PCR array (SABiosciences) was used. This allowed the expression profiling of 84 genes key to cell cycle regulation, including genes whose products both positively and negatively regulate the cell cycle, the transitions between each of the phases, DNA replication, checkpoints, and arrest. Forty-five mRNAs showed a 2-fold or greater change in abundance in HRSV-infected cells, and these are listed in Table 1. For several mRNAs, there was a close correlation between changes in abundance in HRSV-infected primary airway epithelial cells and changes in the corresponding protein abundance detected in a recent quantitative proteomic analysis of A549 cells infected with HRSV subgroup A or B (28) viruses (Table 1, shaded rows). In general, if the abundance of an mRNA was decreased more than 2-fold, this was also reflected in a decrease in the abundance of the corresponding protein (Table 1). No change was observed in the abundance of the mRNA encoding the tumor suppressor protein p53 in HRSV-infected cells, but decreases were found for the mRNAs encoding cyclin D2 and CDK4.
Table 1.
Regulation of mRNAs encoding cell cycle-related proteins at 24 h postinfection in HRSV-infected primary airway epithelial cellsa
| Cell cycle phase or function and gene name | Proteinb | Function, interaction, and regulation in HRSV-infected cells | Fold change in mRNA level in HRSV-infected cells |
|---|---|---|---|
| G1 phase and G1/S transition | |||
| CCND2 | Cyclin D2 | Essential for the control of the cell cycle at the G1/S transition; interacts with CDK4 and CDK6 | −9.1 |
| CCNE1 | Cyclin E1 | Essential for the control of the cell cycle at the G1/S transition; interacts with CDK2; low levels reported in bronchial epithelial cells, consistent with terminal differentiation (40) | −2.17 |
| CDK4 | Cyclin-dependent kinase 4 | Interacts with cyclin D1 | −7.2 |
| CDKN1B | Cyclin-dependent kinase inhibitor 1B (p27, Kip1) | Involved in G1 arrest; inhibitor of cyclin E and cyclin A-CDK2 complexes | 2.11 |
| CDKN3 | Cyclin-dependent kinase inhibitor 3 | Dephosphorylates CDK2 | −6.72 |
| CUL3 | Cullin 3 | Involved in ubiquitinylation of cyclins D1 and E, and hence involved in regulation of the G1/S transition | −2.07 |
| SKP2 | S-phase kinase-associated protein 2 (p45) | Specifically recognizes phosphorylated CDKN1B/p27/Kip1; involved in regulation of the G1/S transition | −3.14 |
| S phase and DNA replication | |||
| MCM3 | Minichromosome maintenance complex component 3 | Allows DNA to undergo a single round of replication per cell cycle | −5.24 |
| MCM4 | Minichromosome maintenance complex component 4 | Involved in the control of DNA replication | −5.09 |
| MCM5 | Minichromosome maintenance complex component 5 | Involved in the control of DNA replication. Quantitative proteomics revealed 9- and 3-fold decreases in the relative abundance of this protein in the nuclei of A549 cells infected with HRSV subgroups A and B, respectively, but no change in the relative abundance in the cytoplasm (27, 28). | −3.45 |
| PCNA | Proliferating cell nuclear antigen | Involved in the control of DNA replication | −4.32 |
| G2 phase and G2/M transition | |||
| BCCIP | BRCA2- and CDKN1A-interacting protein | May promote cell cycle arrest by enhancing the inhibition of CDK2 activity by CDKN1A | −2.15 |
| BIRC5 | Baculoviral IAP repeat-containing 5 | Part of the chromosomal passenger complex that acts as a key regulator of mitosis | −8.88 |
| CCNB1 | Cyclin B1 | Control of the cell cycle at the G2/M transition | −4.83 |
| CDK7 | Cyclin-dependent kinase 7 | Cyclin-activating kinase; activates the cyclin-associated kinases CDK1, CDK2, CDK4, and CDK6 by threonine phosphorylation | 2.33 |
| DDX11 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 11 (CHL1-like helicase homolog, Saccharomyces cerevisiae) | DNA helicase involved in cellular proliferation and expressed in dividing cells | −2.98 |
| GTSE1 | G2 and S-phase expressed 1 | mRNA expressed in the G2/M phase; protein overexpression delays the G2/M phase progression | −4.95 |
| KPNA2 | Karyopherin α2 (RAG cohort 1, importin α1) | Involved in nuclear import. Quantitative proteomics revealed a 7-fold decrease in the abundance of this protein in the nuclear fractions of A549 cells infected with HRSV subgroup A (27). | −4.48 |
| SERTAD1 | SERTA domain containing 1 | Stimulates E2F-1/DP-1 transcriptional activity | 3.08 |
| M phase | |||
| CCNB2 | Cyclin B2 | Control of the cell cycle at the G2/M transition | −5.49 |
| CDC20 | Cell division cycle 20 homolog (S. cerevisiae) | Required for full ubiquitin ligase activity of the anaphase-promoting complex/cyclosome (APC/C); synthesis is initiated at G1/S | −8.11 |
| MRE11A | Meiotic recombination 11 homolog A (S. cerevisiae) | Role in maintenance of telomere integrity | −7.01 |
| RAD51 | RAD51 homolog (S. cerevisiae) (RecA homolog, Escherichia coli) | Involved in the response to DNA damage; interacts with p53 | −3.81 |
| Cell cycle checkpoint and arrest | |||
| ATR | Ataxia telangiectasia and Rad3 related | Serine/threonine kinase that activates the checkpoint; promotion of DBA repair and apoptosis | −2.12 |
| CDC2 | Cell division cycle 2, G1 to S and G2 to M | Quantitative proteomics revealed a 5-fold decrease in the relative abundance of this protein in the nuclei of A549 cells infected with HRSV subgroups A and B (27, 28). | −4.22 |
| CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21WAF1/CIP1) | Binds to and inhibits cyclin-dependent kinase activity, preventing phosphorylation of critical cyclin-dependent kinase substrates and blocking cell cycle progression | 3.07 |
| CDKN2B | Cyclin-dependent kinase inhibitor 2B (p15; inhibits CDK4) | Inhibitor of cyclins | 3.44 |
| GAD | Minichromosome maintenance complex component 2 (MCM) | Required for entry into S phase; allows DNA to undergo a single round of replication per cell cycle | −7.67 |
| GADD45A | Growth arrest and DNA damage inducible, alpha subunit | Binds to PCNA and can inhibit entry of cells into S phase | 5.35 |
| KNTC1 | Kinetochore associated 1 | Part of the mitotic checkpoint; prevents cells from prematurely exiting mitosis | −3.32 |
| MAD2L2 | Mitotic arrest deficient 2-like 2 (yeast) | Involved in mitosis | −3.06 |
| RAD1 | RAD1 homolog (Schizosaccharomyces pombe) | Involved in the cell cycle checkpoint response and DNA repair | −3.34 |
| RAD9A | RAD9 homolog A (S. pombe) | Involved in the cell cycle checkpoint response and DNA repair | 2.38 |
| TFDP1 | Transcription factor Dp-1 | Can stimulate E2F-dependent transcription. The DP2/E2F complex functions in the control of cell cycle progression from G1 to S phase. | −3.54 |
| Negative regulation of the cell cycle | |||
| ATM | Ataxia telangiectasia mutated | Serine/threonine kinase that activates checkpoint signaling upon double-strand breaks; phosphorylates p53 | −4.12 |
| BRCA1 | Breast cancer 1, early onset | DNA-dependent ATPase and 5′-to-3′ DNA helicase required for the maintenance of chromosomal stability | −7.33 |
| RBL1 | Retinoblastoma-like 1 (p107) | Regulates entry into cell division | −3.63 |
| RBL2 | Retinoblastoma-like 2 (p130) | Regulates entry into cell division | −2.03 |
| Other cell cycle regulators | |||
| DIRAS3 | DIRAS family, GTP-binding RAS-like 3 | Present in the cell membrane | 5.6 |
| CCNF | Cyclin F | Acts as an inhibitor of centrosome reduplication | −5.01 |
| CDK5R1 | Cyclin-dependent kinase 5, regulatory subunit 1 (p35) | Neuron-specific activator of CDK5 | 3.99 |
| CKS1B | CDC28 protein kinase regulatory subunit 1B | Binds to the catalytic subunit of the cyclin-dependent kinases and is essential for their biological function | −3.27 |
| CKS2 | CDC28 protein kinase regulatory subunit 2 | Binds to the catalytic subunit of the cyclin-dependent kinases and is essential for their biological function- | −3.03 |
| MKI67 | Antigen identified by monoclonal antibody Ki-67 | Maintains cell proliferation; expression occurs during late G1; cannot be detected in G0 | −5.86 |
| Control mRNAs for PCR | |||
| B2M | β2 microglobulin | PCR control. Quantitative proteomics revealed a 2-fold increase in abundance in the cytoplasm in HRSV subgroup A- and B-infected cells (27, 28). | 12.89 |
| HPRT1 | Hypoxanthine phosphoribosyltransferase 1 | PCR control. Quantitative proteomics revealed no change in the relative abundance of this protein in the nuclear fractions of A549 cells infected with HRSV subgroup A or B (27, 28). | −1.93 |
| RPL13A | Ribosomal protein L13a | PCR control.- | −5.08 |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | PCR control. Quantitative proteomics revealed no change in the relative abundance of this protein in the nuclear fractions of A549 cells infected with HRSV subgroup A or B, a finding also reflected in independent Western blot analysis (27, 28). | −1.2 |
| ACTB | Actin, beta | PCR control | −1.1 |
Protein functions are annotated based on previous work on HRSV. In cases where the relative abundance of the protein encoded by the mRNA has been shown by quantitative proteomics using stable isotope labeling with amino acids in cell culture (SILAC) (27, 28) to be changed in A549 cells infected with HRSV subgroups A and B, the row is shaded.
IAP, inhibitor of apoptosis proteins; RAG, recombinase-activating gene.
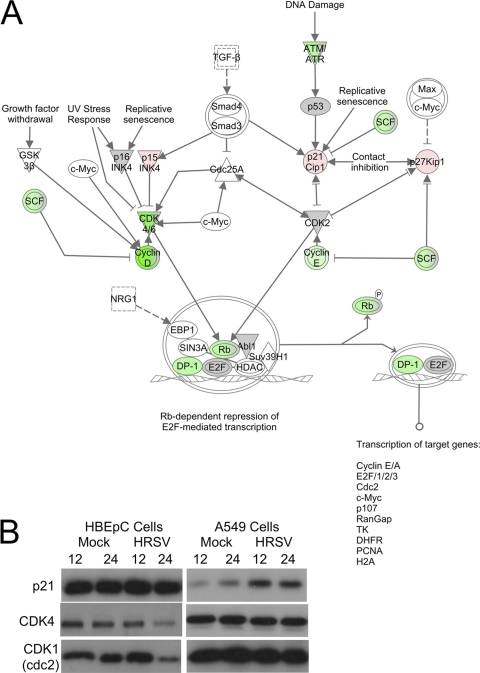
Control of the cell cycle is a complex, multilayered process. Network pathway analysis of the transcriptomic data set using Ingenuity Pathway Analysis indicated that the top canonical pathways affected would be cell cycle regulation (P, 1.11 × 10−21), and specifically G1/S phase checkpoint regulation (P, 9.77 × 10−20) (Fig. 2A). Immunoblot analysis of the abundances of p53, p21WAF1/CIP1, and CDK4 proteins in HRSV-infected HBEpC cells showed no change in p53 (data not shown) or p21WAF1/CIP1 but a decrease in the abundance of CDK4 protein from that in mock-treated cells at 24 h (Fig. 2B), reflecting the transcriptomic data set. This was in contrast to HRSV-infected A549 cells, which showed an increase in the abundance of p21WAF1/CIP1 protein over that in mock-treated cells at 12 and 24 h (Fig. 2B).
Fig. 2.
(A) Ingenuity Pathway Analysis of the transcriptomic data sets, showing the relationship between cell cycle-regulatory factors whose mRNAs either were decreased in abundance more than 2-fold (green), remained unchanged (gray), or increased in abundance more than 2-fold (red) in HRSV-infected HBEpC cells. This diagram focuses on molecules involved in the G1/S phase transition. (B) Immunoblot analysis of the abundances of p21WAF1/CIP1 (p21), CDK4, and CDK1 at 12 and 24 h postinfection in HBEpC and A549 cells that were either mock treated or infected with HRSV.
Similarly, a decrease in the level of the mRNA encoding the cell cycle-regulatory transcription factor DP-1 (TFDP-1, which complexes with E2F family members) would be predicted to lead to decreases in the expression of several target genes related to the regulation of the cell cycle, which again is observed in this study, including a decrease in the abundance of CDK1 mRNA (Table 1). This was also confirmed by immunoblot analysis of HRSV-infected HBEpC cells, which showed a decrease in the abundance of CDK1 protein (Fig. 2B), but this decrease was not observed in HRSV-infected A549 cells (Fig. 2B).
HRSV-infection resulted in G0/G1 phase enrichment in both HBEpC and A549 cells.
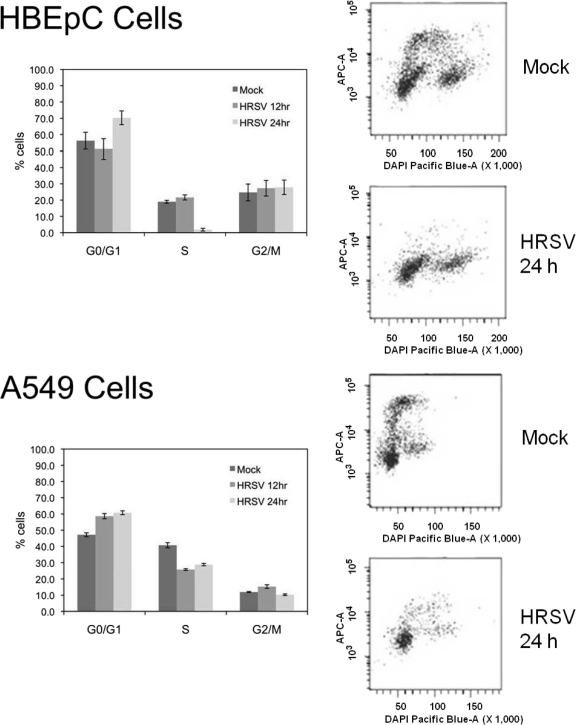
The decreased abundances of cyclin D1, cyclin D2, CDK4, and CDK1 proteins in HRSV-infected HBEpC cells and the increase in the abundance of p21WAF1/CIP1 protein in A549 cells would both be predicted to result in an accumulation of infected cells in the G0/G1 phase of the cell cycle in the respective cell types. The D-type cyclins and the CDK4 and CDK1 proteins are all required for progression from the G1 phase into the S phase of the cell cycle, and decreases in the abundances of these proteins would result in an arrest in the G0/G1 phase of the cell cycle. p21WAF1/CIP1 is a CDK inhibitor; it binds to CDK4 or CDK2 complexes and thus inhibits the activities of the cyclin D–CDK4 and cyclin E–CDK2 complexes (16, 37). To test whether G0/G1 phase arrest occurred in HRSV-infected cells, the proportion of cells in each stage of the cell cycle was determined, by use of dual-label flow cytometry, in both mock-treated and HRSV-infected HBEpC and A549 cells. In this approach, DAPI was used to stain the nucleus (from which the proportions of cells in the G0/G1 and G2/M phases were derived), and EdU was added and incorporated into replicating DNA (from which the proportion of cells in the S phase was derived). In both cell types, HRSV infection resulted in an enrichment of cells in the G1 phase of the cell cycle and a reduction in the proportion of cells in the S phase (Fig. 3).
Fig. 3.
Cell cycle profiles for HBEpC and A549 cells dually stained with EdU and DAPI. (Left) Bars in histograms represent the mean percentages of cells in the G0/G1, S, and G2/M phases of the cell cycle in mock-treated and HRSV-infected cells at 12 and 24 h postinfection. (Right) Representative cell cycle profiles of dually stained (EdU/DAPI) mock-treated and HRSV-infected (24-h) HBEpC and A549 cells are shown for each experimental condition.
Blocking G1 phase progression resulted in an increase in progeny virus production.
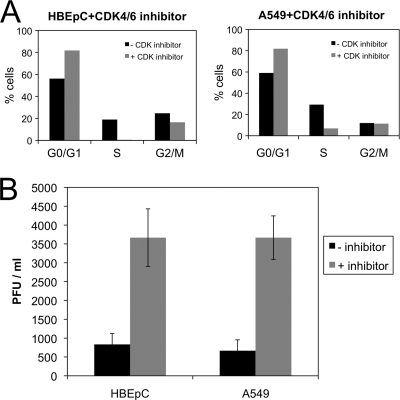
In general, with viruses that cause aberrations in the cell cycle, the stage where a cell cycle delay occurs can also be advantageous for progeny virus production (5). To investigate whether an arrest in the G0/G1 phase was advantageous for HRSV progeny virus production, both HBEpC and A549 cells were treated 1 h prior to infection with a kinase inhibitor specific to CDK4/6 proteins and were maintained in this inhibitor throughout infection. The inhibitor prevented the activity of the cyclin D–CDK4/6 complex in promoting the progression of the cell cycle through the G0/G1 phase. The proportions of cells in the different cell cycle stages after 24 h were determined by dual-label flow cytometry and were compared to those for cells grown in the absence of inhibitor. The data indicated that in the presence of the inhibitor, cells accumulated in the G0/G1 phase of the cell cycle (Fig. 4A). The effects of the absence and presence of the inhibitor on virus progeny production were determined by a plaque assay. In the presence of the inhibitor, the data indicated an approximately 5-fold increase in the number of plaques for both cell types (Fig. 4B). This indicated that the G0/G1 phase of the cell cycle was preferential for progeny virus production compared to asynchronously growing cells.
Fig. 4.
(A) Histograms showing the proportion of cells in each stage of the cell cycle for HBEpC and A549 cells grown in the absence (−) or presence (+) of the specific inhibitor of CDK4/6. (B) Change in the amount of output virus from HRSV-infected HBEpC or A549 cells grown in the absence (−) or presence (+) of the specific inhibitor of CDK4/6, as determined by a plaque assay. These experiments were performed in triplicate and are representative of technical replicates.
DISCUSSION
Several different viruses with RNA genomes have been shown to interact with the cell cycle to create conditions inside the cell that promote progeny virus production (4, 8, 11, 19, 22, 23, 29, 34). The interaction of HRSV with the cell cycle has been studied previously in both a primary cell type and continuous cell culture (A549 and HEp-2 cells) (12, 26). Although these studies demonstrated that cell cycle arrest occurred in HRSV-infected cells and was beneficial for progeny virus production, potential changes to cell cycle-regulatory complexes involved in cell cycle modulation were not extensively investigated. However, the stage of cell cycle arrest was found to differ in the different cell types. In the current study, the abundances of cell cycle-regulatory factors and corresponding mRNAs were investigated in the context of primary cells and also, for comparison, in A549 cells (where appropriate).
Several differences in the abundances of cell cycle-regulatory molecules were observed between HRSV-infected HBEpC and A549 cells, on the one hand, and mock-treated cells, on the other (Fig. 1 and 2). The abundances of cyclin D1, cyclin D2, CDK4, and CDK1 proteins decreased in HRSV-infected HBEpC cells but not in HRSV-infected A549 cells, where instead the abundance of p21WAF1/CIP1 protein increased. This study presented the first targeted analysis of the cell cycle transcriptome in HRSV-infected cells (which were primary cells). Where previous studies have investigated mRNA levels in HRSV-infected cells, which included mRNAs encoding cell cycle-regulatory proteins, there was close agreement between these and the current study. For example, in this study, the mRNA encoding cyclin D1 did not change more than 2-fold in abundance. Similarly, the abundances of mRNAs encoding cyclin D1 and cyclin G2 (an unconventional cyclin that blocks cell cycle entry and is highly expressed in postmitotic cells) did not change more than 2-fold in A549 cells infected with either the A2 or the Long strain of HRSV (25). Our findings, taken with those of other studies (25), indicate that there is no general shutoff of host cell transcription in HRSV-infected airway epithelial cells. In general, the level of an mRNA in HRSV-infected cells compared to that in mock-treated cells also corresponded to the level of the protein as determined by this study or by quantitative proteomic analysis (27, 28).
D-type cyclin (D1 and D2) proteins can act differently and have different activities; for example, in primary cardiomyocytes, cyclin D2 protein was much less effective at activating CDK2 protein and cell proliferation than cyclin D1 protein (38). Apart from abundance, the activities of D-type cyclin proteins can also be regulated by their localization (17). In mock-treated cells, cyclin D1 and D2 proteins were predominately nuclear (Fig. 1C). In contrast, in HRSV-infected cells, the localization of these cyclins was also cytoplasmic. There was no obvious change in the subcellular localization of CDK1 protein (Fig. 1D), indicating that changes in localization were specific to cyclin D1 and D2 proteins and did not reflect a general redistribution of cellular proteins between the nucleus and the cytoplasm in HRSV-infected cells. This was also reflected in quantitative proteomic analysis of the nuclear and cytoplasmic proteomes in HRSV-infected A549 cells, where no large-scale redistribution of proteins was observed and changes in localization were confined to proteins associated with specific pathways and networks (27, 28).
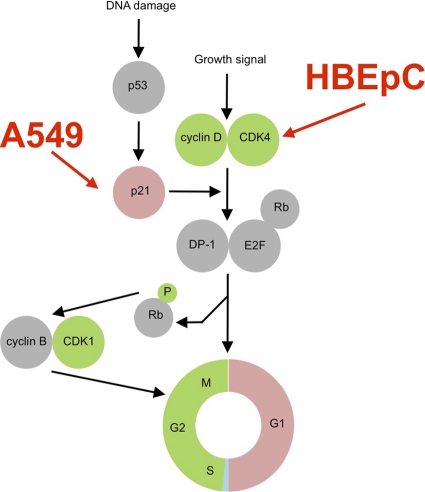
Interestingly, the changes in the abundances of cell cycle-regulatory molecules in HRSV-infected cells, mainly the decreases in the levels of D-type cyclin proteins and CDK4 proteins in HBEpC cells and the increased abundance of p21WAF1/CIP1 protein in A549 cells, were predicted to have the same cell cycle outcome—an enrichment of infected cells in the G0/G1 phase (Fig. 5). This was confirmed by dual-label flow cytometry analysis of the cell cycle stages in HRSV-infected cells compared to mock-treated cells, which indicated enrichment in the G0/G1 phase population in both HBEpC and A549 cells infected with HRSV. This stage of the cell cycle also correlated with increased progeny virus production compared to that by virus grown in asynchronously growing cells (Fig. 4). Enrichment rather than a total arrest was observed, probably because investigation of cell cycle phenotypes much beyond 24 h in this type of study is problematic due to the effects of virus infection on cells (mainly apoptosis and cell death). In conventional cell cycle analysis, cells are often grown for as long as 72 h in order for cell cycle aberrations to become manifest, i.e., to allow several cell cycles and the resulting accumulation of cells in one particular phase. In addition, the starting amount of virus and the proportion of cells initially infected, which again may be cell cycle related, will determine the proportion of cells showing cell cycle changes. Although the use of primary cells is advantageous, they are a mixed population; therefore, not all cells will be susceptible to infection, and the pharmacological background and nature of the starting population of cells are generally unknown.
Fig. 5.
Model showing examples of cell cycle-regulatory proteins involved in progression whose abundances either increased (red) or decreased (green) in HRSV-infected cells. The model shows that although the abundances of different proteins were affected differently in HRSV-infected HBEpC or A549 cells, these changes will still result in G0/G1 phase arrest. In addition, a reduction in the abundance of CDK1 is observed in HRSV-infected cells at 24 h postinfection, and this may act on G2/M phase progression.
The current study presented results different from those of a previous analysis of HRSV-infected primary and continuous (A549) cells (12). In both studies, A549 cells infected with HRSV were enriched in the G0/G1 phase, whereas in the current study, HRSV-infected primary cells were also enriched in the G0/G1 phase, in contrast with an arrest in the G2/M phase in this cell type (12). The results of cell cycle analysis were equivalent in that dual-label flow cytometry was used to accurately determine the proportion of cells in each stage, and differences in these proportions may reflect differences in primary cells and growth conditions. Although the primary airway epithelial cells used in the current study represent in vivo targets of infection, they are again different from well-differentiated primary bronchial epithelial cells (32, 40), which may, again, have different cell cycle profiles.
The interaction of HRSV with the cell cycle and the possible remodeling of airway epithelial cells may be a causative pathway to disease (12), and general respiratory virus-induced cell cycle dysfunction may be a contributing factor to changes in airway development. The pharmacological inhibition of the cell cycle in G0/G1 phase and the resulting increase in the efficiency of progeny virus production over that for HRSV grown in asynchronously replicating cells also have potential implications for vaccinology in that manipulation of cell growth conditions can lead to more-efficient virus production. How the arrest in the G0/G1 phase is beneficial to HRSV biology remains to be elucidated. Viruses may arrest or delay cells in the cell cycle prior to mitosis in order to prevent the reorganization of the endoplasmic reticulum and the Golgi apparatus that occurs during nuclear division and cytokinesis—and thus interferes with virus replication and assembly complexes (23)—or because in the G1 phase of the cell cycle, translation is maximal.
ACKNOWLEDGMENTS
J.N.B. is a Research Council UK Fellow, and J.A.H. is a holder of a Leverhulme Trust Research Fellowship. D.C.M. is the recipient of an MRC Ph.D. studentship.
Footnotes
Published ahead of print on 27 July 2011.
REFERENCES
- 1. Belzile J. P., Abrahamyan L. G., Gerard F. C., Rougeau N., Cohen E. A. 2010. Formation of mobile chromatin-associated nuclear foci containing HIV-1 Vpr and VPRBP is critical for the induction of G2 cell cycle arrest. PLoS Pathog. 6:e1001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bruni R., Roizman B. 1998. Herpes simplex virus 1 regulatory protein ICP22 interacts with a new cell cycle-regulated factor and accumulates in a cell cycle-dependent fashion in infected cells. J. Virol. 72:8525–8531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cawood R., Harrison S. M., Dove B. K., Reed M. L., Hiscox J. A. 2007. Cell cycle dependent nucleolar localization of the coronavirus nucleocapsid protein. Cell Cycle 6:863–867 [DOI] [PubMed] [Google Scholar]
- 4. Chen C. J., Makino S. 2004. Murine coronavirus replication induces cell cycle arrest in G0/G1 phase. J. Virol. 78:5658–5669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Davy C., Doorbar J. 2007. G2/M cell cycle arrest in the life cycle of viruses. Virology 368:219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Diehl J. A., Cheng M., Roussel M. F., Sherr C. J. 1998. Glycogen synthase kinase-3β regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12:3499–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Diehl J. A., Zindy F., Sherr C. J. 1997. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 11:957–972 [DOI] [PubMed] [Google Scholar]
- 8. Dove B., Brooks G., Bicknell K., Wurm T., Hiscox J. A. 2006. Cell cycle perturbations induced by infection with the coronavirus infectious bronchitis virus and their effect on virus replication. J. Virol. 80:4147–4156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Eckardt-Michel J., et al. 2008. The fusion protein of respiratory syncytial virus triggers p53-dependent apoptosis. J. Virol. 82:3236–3249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Engelking O., Fedorov L., Lilischkis R., ter Meulen V., Schneider-Schaulies S. 1999. Measles virus-induced immunosuppression in vitro is associated with deregulation of G1 cell cycle control proteins. J. Gen. Virol. 80:1599–1608 [DOI] [PubMed] [Google Scholar]
- 11. Feuer R., Mena I., Pagarigan R., Slifka M. K., Whitton J. L. 2002. Cell cycle status affects coxsackievirus replication, persistence, and reactivation in vitro. J. Virol. 76:4430–4440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gibbs J. D., Ornoff D. M., Igo H. A., Zeng J. Y., Imani F. 2009. Cell cycle arrest by transforming growth factor β1 enhances replication of respiratory syncytial virus in lung epithelial cells. J. Virol. 83:12424–12431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Groschel B., Bushman F. 2005. Cell cycle arrest in G2/M promotes early steps of infection by human immunodeficiency virus. J. Virol. 79:5695–5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Groskreutz D. J., et al. 2007. Respiratory syncytial virus decreases p53 protein to prolong survival of airway epithelial cells. J. Immunol. 179:2741–2747 [DOI] [PubMed] [Google Scholar]
- 15. Guo Y., et al. 2005. Phosphorylation of cyclin D1 at Thr 286 during S phase leads to its proteasomal degradation and allows efficient DNA synthesis. Oncogene 24:2599–2612 [DOI] [PubMed] [Google Scholar]
- 16. Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. 1993. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75:805–816 [DOI] [PubMed] [Google Scholar]
- 17. Harrison S. M., et al. 2007. Characterisation of cyclin D1 down-regulation in coronavirus infected cells. FEBS Lett. 581:1275–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. He J., et al. 1995. Human immunodeficiency virus type 1 protein R (Vpr) blocks cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705–6711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. He Y., et al. 2010. Influenza A virus replication induces cell cycle arrest in G0/G1 phase. J. Virol. 84:12832–12840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Izumi T., et al. 2010. HIV-1 viral infectivity factor interacts with TP53 to induce G2 cell cycle arrest and positively regulate viral replication. Proc. Natl. Acad. Sci. U. S. A. 107:20798–20803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laine D., et al. 2005. Measles virus nucleoprotein induces cell-proliferation arrest and apoptosis through NTAIL-NR and NCORE-FcγRIIB1 interactions, respectively. J. Gen. Virol. 86:1771–1784 [DOI] [PubMed] [Google Scholar]
- 22. Li F. Q., Tam J. P., Liu D. X. 2007. Cell cycle arrest and apoptosis induced by the coronavirus infectious bronchitis virus in the absence of p53. Virology 365:435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin G. Y., Lamb R. A. 2000. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J. Virol. 74:9152–9166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lomonte P., Everett R. D. 1999. Herpes simplex virus type 1 immediate-early protein Vmw110 inhibits progression of cells through mitosis and from G1 into S phase of the cell cycle. J. Virol. 73:9456–9467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martinez I., Lombardia L., Garcia-Barreno B., Dominguez O., Melero J. A. 2007. Distinct gene subsets are induced at different time points after human respiratory syncytial virus infection of A549 cells. J. Gen. Virol. 88:570–581 [DOI] [PubMed] [Google Scholar]
- 26. Mohapatra S., et al. 2009. Human respiratory syncytial virus reduces the number of cells in S-phase and increases GADD153 expression in HEp-2 cells. Acta Virol. 53:207–211 [DOI] [PubMed] [Google Scholar]
- 27. Munday D., et al. 2010. Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus. Mol. Cell. Proteomics 9:2438–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Munday D., Hiscox J. A., Barr J. N. 2010. Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus subgroup B using stable isotope labeling with amino acids in cell culture (SILAC) coupled to LC-MS/MS. Proteomics 10:4320–4334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Naniche D., Reed S. I., Oldstone M. B. A. 1999. Cell cycle arrest during measles virus infection: a G0-like block leads to suppression of retinoblastoma protein expression. J. Virol. 73:1894–1901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Niewiesk S., et al. 1999. Measles virus-induced immunosuppression in cotton rats is associated with cell cycle retardation in uninfected lymphocytes. J. Gen. Virol. 80:2023–2029 [DOI] [PubMed] [Google Scholar]
- 31. Ojala P. M., et al. 2000. The apoptotic v-cyclin-CDK6 complex phosphorylates and inactivates Bcl-2. Nat. Cell Biol. 2:819–825 [DOI] [PubMed] [Google Scholar]
- 32. Parker J., et al. 2010. A 3-D well-differentiated model of pediatric bronchial epithelium demonstrates unstimulated morphological differences between asthmatic and nonasthmatic cells. Pediatr. Res. 67:17–22 [DOI] [PubMed] [Google Scholar]
- 33. Parnell G., et al. 2011. Aberrant cell cycle and apoptotic changes characterise severe influenza A infection—a meta-analysis of genomic signatures in circulating leukocytes. PLoS One 6:e17186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Poggioli G. J., Keefer C., Connolly J. L., Dermody T. S., Tyler K. L. 2000. Reovirus-induced G2/M cell cycle arrest requires σ1s and occurs in the absence of apoptosis. J. Virol. 74:9562–9570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Re F., Braaten D., Franke E. K., Luban J. 1995. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J. Virol. 69:6859–6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schnorr J., et al. 1997. Cell cycle arrest rather than apoptosis is associated with measles virus contact-mediated immunosuppression in vitro. J. Gen. Virol. 78:3217–3226 [DOI] [PubMed] [Google Scholar]
- 37. Stacey D. W. 2003. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr. Opin. Cell Biol. 15:158–163 [DOI] [PubMed] [Google Scholar]
- 38. Tamamori-Adachi M., Goto I., Yamada K., Kitajima S. 2008. Differential regulation of cyclin D1 and D2 in protecting against cardiomyocyte proliferation. Cell Cycle 7:3768–3774 [DOI] [PubMed] [Google Scholar]
- 39. Upton J. W., van Dyk L. F., Speck S. H. 2005. Characterization of murine gammaherpesvirus 68 v-cyclin interactions with cellular cdks. Virology 341:271–283 [DOI] [PubMed] [Google Scholar]
- 40. Villenave R., et al. 2010. Cytopathogenesis of Sendai virus in well-differentiated primary pediatric bronchial epithelial cells. J. Virol. 84:11718–11728 [DOI] [PMC free article] [PubMed] [Google Scholar]