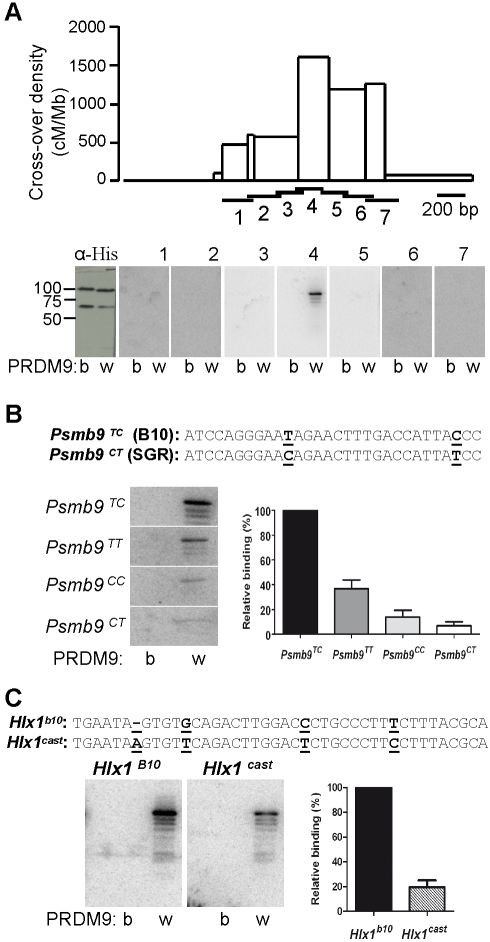
Figure 2. PRDM9 binds in vitro to meiotic recombination hotspots.
(A) Detection by southwestern blotting of PRDM9 binding at the Psmb9 recombination hotspot. Upper panel, CO distribution along the Psmb9 hotspot [39]. Horizontal bars show the positions of the DNA probes (numbered from 1 to 7) used for southwestern experiments. Lower panel, PRDM9 (b, His-PRDM9b; w, His-PRDM9wm7) was probed with anti-His antibody and the radio-labeled double-stranded DNA probes 1–7 (about 200 bp). The molecular weights of His-PRDM9b and His-PRDM9wm7 are 101 kDa and 98 kDa, respectively. The bands with lower molecular weights correspond to PRDM9 degradation products. (B) Effect of SNPs at the center of the Psmb9 hotspot on the in vitro binding of PRDM9. The sequence of the likely PRDM9wm7 binding sequence [13] is shown, and the SNPs between the B10 and B10.MOL-SGR strains are underlined (see FigS5 for in silico prediction). PRDM9b and PRDM9wm7 were probed with radio-labeled double-stranded oligonucleotides that carried the four possible SNP combinations (60 bp, Table S14). The amount of signal due to binding of each probe to PRDM9wm7 is shown (with standard error), relative to Psmb9TC. The decrease of binding to the double mutant probe Psmb9 CT (0.07% binding relative to Psmb9 TC) is consistent with a cumulative effect of each single mutant (0.36% and 0.14% binding relative to Psmb9 TC) suggesting their effects are independent. (C) Analysis by southwestern blotting of PRDM9 binding to the putative PRDM9wm7 binding motif at the center of the Hlx1 hotspot [13]. The likely PRDM9wm7 binding sequences in the B10 and CAST strains are shown, with SNPs underlined (see Figure S5 for in silico prediction). PRDM9b and PRDM9wm7 were probed with radio-labeled double-stranded oligonucleotides that carried B10 or CAST allele (41 bp, Table S14). Signal intensities of the binding of the Hlx1B10 and Hlx1cast probes (relative to HlxB10) to PRDM9wm7 are shown.