Figure 3. Neighbour-Joining phylogenetic trees.
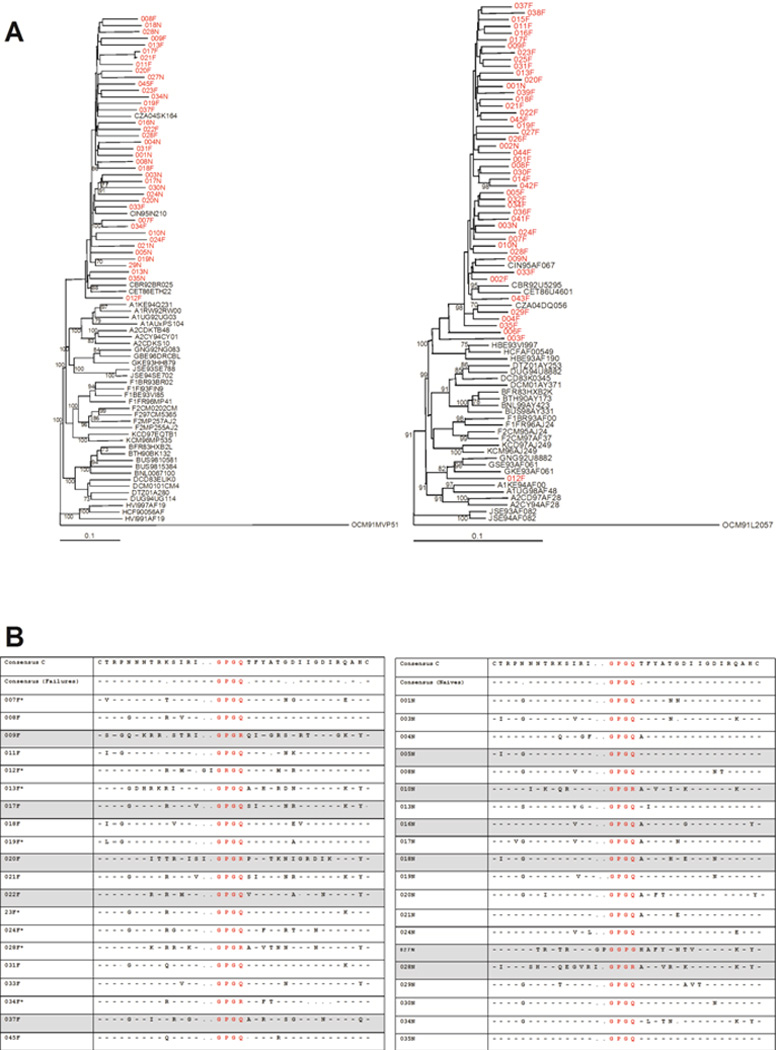
A) Left panel - Neighbour-Joining phylogenetic tree constructed from the env gene sequences. All clones highlighted in red cluster closely with subtype C. Right panel - Neighbour-Joining phylogenetic tree constructed from the pol gene sequences. All patient samples highlighted in red cluster together with the subtype C reference with the exception of 704MC012F which clusters with subtype G. Boostrap values more than 70% are shown. B) Alignment of V3 sequences of clones of ARV-failing and ARV-naïve patients. The panel on the left indicates V3 sequences in patients failing treatment and the panel on the right indicates V3 sequences in ARV-naive patients. The crown motif for each sequence is highlighted in red. All sequences from viruses determined to be X4/dual/mixed by the Trofile assay are highlighted in grey and samples with asterisk next to sample number were not available for analysis in the Trofile assay or did not yield reportable data.
