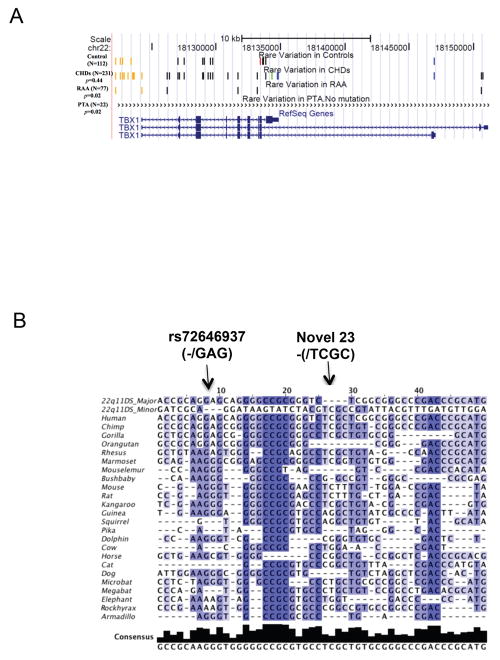
Figure 4.
(A) Enrichment of very rare DNA variations. “Control”, “ CHDs” (Congenital heart defects=CTDs+ASD), “RAA” and “PTA” custom tracks were added to the UCSC Genome Browser as shown. The custom tracks indicate the type of rare variations (MAF<0.01) identified by TBX1 resequencing analysis in each phenotype enrichment group (total number of subjects and definition of each phenotype group is in Figure 1). Each line or bar represents a different rare SNP. The orange vertical lines represent variations from the putative promoter region, black represents intronic variants and blue are variants from the 3′-UTR. The red bar represents a nonsynonymous variant and a green bar denotes a synonymous variation. Detailed information of rare variations used for DNA enrichment analysis is shown in Supp. Table S2. (B) Multiple sequence alignment of 45 very rare TBX1 variants (MAF<0.01). Multiple alignment data from 44 vertebrate species generated using multiz and other tools in the UCSC/Penn State Bioinformatics comparative genomics alignment pipeline were extracted using Galaxy (http://main.g2.bx.psu.edu/), a web-based genome analysis tool (Blankenberg, et al., 2010; Goecks, et al., 2010). Both Galaxy and then Jalview (Waterhouse, et al., 2009) were used to edit alignments. The species in the database with significant missing sequence were discarded and only results from 24 mammals are shown. The first two rows indicate the major or minor allele identified in the 22q11DS patient cohort. Each row below represents one of the 24 mammalian species. In general, each column represents a rare DNA variation except for the 8th variation that has a three base and 24th that has a four base indel. The most intense blue color corresponds to the most conserved SNPs. A total of 30 major alleles from the 22q11DS cohort show evolutionary conservation. Five minor alleles show conservation as indicated. The minor allele (TCGC), the only novel variation (novel 23) found in the sample, was conserved among most mammalian species. Detailed information on the 45 rare variations is shown in Supp. Table S2.