Figure 6.
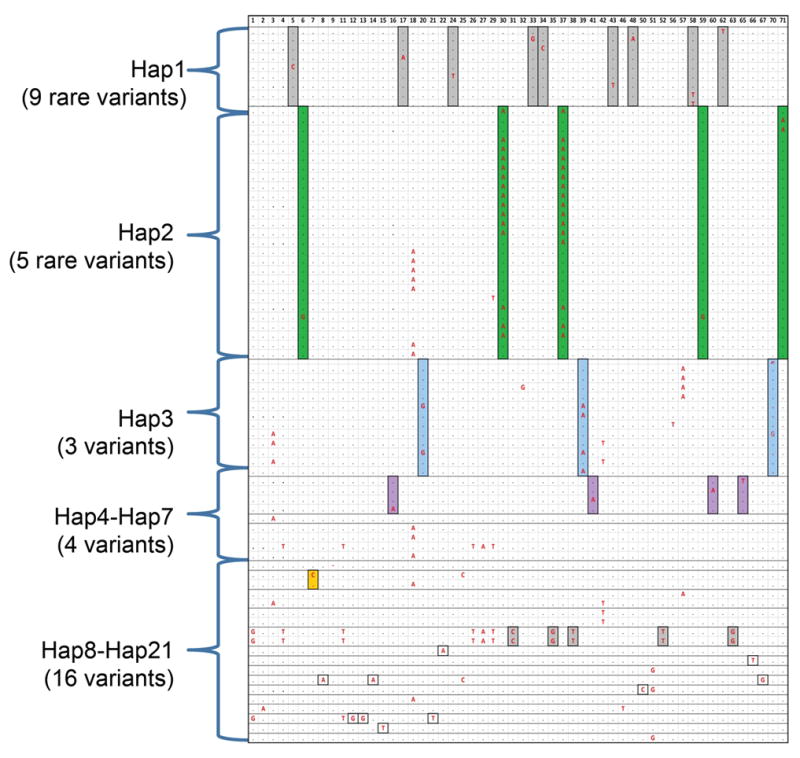
Haplotypes of common SNPs (MAF>0.05) and 54 rare variants (MAF<0.05). A total of 54 rare variants (MAF<0.05) identified by TBX1 resequencing were shown in each column (the order of variants is according to Supp. Table S2). Each row represents a patient that carries one or more of the 54 rare variants (the major allele is a black dot; the minor allele is indicted in bold red font). Each haplotype group is indicated as Hap1–21 (Hap22–49 which didn’t occur in any patient with rare variations, is not shown.). They were determined by the allele pattern of the 17 common SNPs. Some patients shared common haplotypes. Twenty-one of 49 haplotypes captured all rare variations (MAF<0.05) in a total of 77 subjects. Hap1 (CAGGGTCTCAAAACGCA), Hap2 (CTCGGTCTCAAAACGCA) and Hap3 (TTCGACCCCGGACCTCG) were common haplotypes (frequency>0.05) generated by 17 common SNPs from TBX1 resequencing data. Hap4 to Hap7 were less common haplotypes (0.01<frequency<0.05) and hap8 to hap21 were very rare haplotypes (frequency<0.01). For each variant, the boxes highlighted in different colors represent the minor allele of rare variants observed in the subjects with the given haplotypes indicated (Hap1–21).
