Abstract
Important questions in biology have emerged recently concerning the timing of transcription in living cells. Studies on clonal cell lines have shown that transcription is often pulsatile and stochastic, with implications for cellular differentiation. Currently, information regarding transcriptional activity at cellular resolution within a physiological context remains limited. To investigate single-cell transcriptional activity in real-time in living tissue we used bioluminescence imaging of pituitary tissue from transgenic rats in which luciferase gene expression is driven by a pituitary hormone gene promoter. We studied fetal and neonatal pituitary tissue to assess whether dynamic patterns of transcription change during tissue development. We show that gene expression in single cells is highly pulsatile at the time endocrine cells first appear but becomes stabilised as the tissue develops in early neonatal life. This stabilised transcription pattern might depend upon tissue architecture or paracrine signalling, as isolated cells, generated from enzymatic dispersion of the tissue, display pulsatile luminescence. Nascent cells in embryonic tissue also showed coordinated transcription activity over short distances further indicating that cellular context is important for transcription activity. Overall, our data show that cells alter their patterns of gene expression according to their context and developmental stage, with important implications for cellular differentiation.
Key words: Prolactin, Transcription, Pituitary, Development, Microscopy
Introduction
Gene expression and the process of transcription is an intensively studied area of cell biology. Quantification of transcription activity is classically performed on populations of cells at set time points providing an averaged snapshot view of the cellular transcription response to the conditions under investigation. In recent years, efforts have been made to quantify transcription activity in single cells and in real-time, which has been facilitated by technical advances in live-cell microscopy and the continued development of reporter proteins (Larson et al., 2009). The use of reporter proteins as a read-out of transcription activity has shown that transcription is often pulsatile and highly variable between cells, even in clonal cell populations (Elowitz et al., 2002; Blake et al., 2003; Raser and O'Shea, 2004). However, currently, there is limited information regarding transcriptional activity at cellular resolution within a physiological context.
The pituitary gland is an ideal system in which to study transcription activity and its regulation. The gland, an essential component of the endocrine system, secretes the hormones prolactin (PRL), growth hormone, thyrotrophin, gonadotrophins and adrenocorticotrophin, which are generated from specific differentiated cells located within the anterior lobe of the pituitary. The production and secretion of these hormones are regulated by numerous molecules. The differentiated cells within the pituitary gland arise at specific time points during pituitary development in response to the action of various signalling molecules and transcription factors (Zhu et al., 2007). Lactotrophs, the last cell type to emerge in the pituitary, require the Pit-1 transcription factor (also known as Pou1f1) for terminal differentiation, as do the somatotroph and thyrotroph cell lineages (Camper et al., 1990; Li et al., 1990). Lactotrophs might be functionally heterogeneous and can be subdivided into three types, depending upon secretory granule size, by electron microscopic analysis (Christian et al., 2007). The relative proportion of these cell types varies depending upon sex and age (Takahashi and Miyatake, 1991). The positioning of the different cell types within the pituitary might be functionally organised. Somatotroph cells form a cell network, linked by adherens junctions, that can propagate calcium signals across the gland (Bonnefont et al., 2005). This organisation arises soon after somatotroph cells emerge in the developing pituitary. Folliculostellate cells might also form a connected cell network (Fauquier et al., 2001), but it remains uncertain whether lactotroph cells are similarly organised.
Quantification of PRL gene expression in real-time in pituitary lactotroph cells previously detected pulsatile transcription activity in cell lines (Takasuka et al., 1998; McFerran et al., 2001) and isolated primary cells (Stirland et al., 2003). To assess the transcription dynamics of the PRL gene within individual cells in intact tissue, and the level of cell-to-cell coordination, transgenic F344 rats with firefly luciferase or destabilised EGFP reporter gene expression controlled by more than 155 kbp of the human PRL gene locus were generated (Semprini et al., 2009). Individual cells showed large changes in transcription levels over 15 hours, with little apparent local cell-to-cell coordination (Harper et al., 2010). Cells had different patterns of transcription activity, although it was unclear whether this was due to functional differences between the cells and whether cells could alter these transcription patterns in different physiological contexts.
In situ hybridisation analyses indicate that transcription activity can change during development (Wijgerde et al., 1995; Newlands et al., 1998; Pare et al., 2009). However, this technique only provides a static representation of the cellular activity at a particular time point, preventing detailed characterisation of transcription dynamics. In this study, we report the characterisation of PRL gene expression in real-time and assess how this changes during pituitary tissue development. Bioluminescence imaging was used to quantify transgene expression in intact living pituitary tissue and enzymatically dispersed cells from embryonic and neonatal PRL-luciferase transgenic rats. We show that transcription of the PRL gene initiates earlier than previously determined, at embryonic day (E) 16.5. Luciferase expression in these nascent lactotroph cells showed that transcription occurred in transient pulses, with coordination of cell activity limited to closely apposed cells. In late embryonic and early neonatal pituitaries, pulsatile patterns of transcription were replaced with stable transcription activity. However, after enzymatic dispersion of postnatal pituitary tissue, isolated cells displayed pulsatile luminescence, implying that the stabilised transcription pattern depends upon tissue architecture or local paracrine signalling. These data indicate that cells alter their dynamic patterns of gene transcription according to their context and developmental stage. This has particular implications for cellular differentiation as the change in transcriptional activity might reflect a transitional state between different cell stages.
Results
Organisation of transcriptionally active lactotroph cells within the developing rat pituitary gland
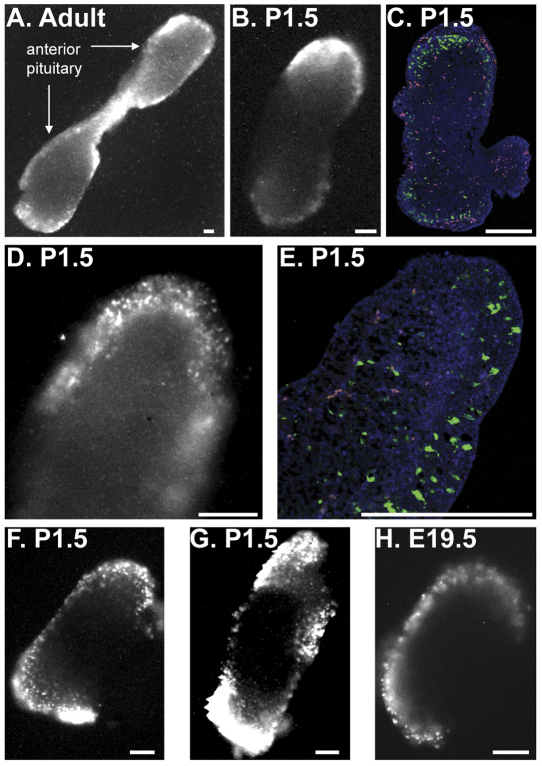
To assess the real-time transcription activity of the PRL gene, transgenic rats were previously generated that express the luciferase reporter gene under the regulation of the human PRL locus (hPRL-Luc49), followed by bioluminescence imaging of cells or tissue in culture in a temperature-, CO2- and humidity-controlled imaging chamber (Semprini et al., 2009; Harper et al., 2010). Characterisation of the reporter transgene showed that it faithfully reflects endogenous Prl gene transcription activity as it is expressed in the correct cell type (lactotroph cells) and responds appropriately to various regulators of Prl expression (Semprini et al., 2009; Harper et al., 2010). The location of transcriptionally active lactotroph cells in the rat pituitary has been assessed by bioluminescence imaging of adult pituitary tissue slices 7. and, in this study, in whole neonatal and embryonic pituitaries from transgenic hPRL-Luc49 rats. Luminescence signal was greatest at the periphery of the pituitary in adult pituitary (Fig. 1A), as noted previously (Harper et al, 2010), in post-natal day (P) 1.5 pituitary (Fig. 1B–G) and E19.5 pituitary (Fig. 1H). Immunofluorescent detection of luciferase protein, following culture and forskolin activation of the pituitaries, also detected a greater number of luciferase-expressing cells at the periphery of the tissue (Fig. 1C,E). This indicates that lactotroph cells with the greatest transcriptional activity are preferentially localised peripherally in the pituitary, and that this topographical organisation of lactotroph cells is established during fetal pituitary development. This organisation might reflect specialisation of lactotroph cells, as functional differences between peripherally and centrally located lactotrophs have been detected previously (Boockfor and Frawley, 1987). Higher magnification of neonatal tissue detected discrete areas of luminescent signal, which appeared to represent single cells and were of a similar size to a typical eukaryotic cell (Fig. 1D,F,G). These areas of luminescent signal were similar in size to those seen in embryonic pituitaries, when lactotroph cells are less abundant, and thus are likely to represent individual cells rather than small cell clusters (Fig. 1G; Fig. 2A). Furthermore, immunofluorescence analysis of P1.5 pituitaries confirmed the clear spatial separation of individual luciferase-expressing cells, comparable to the luminescence images.
Fig. 1.
Organisation of transcriptionally active lactotroph cells within the developing pituitary. Bioluminescence imaging of adult pituitary slices and whole embryonic and neonatal pituitaries. (A) Luminescence image of a 400-μm-thick coronal-orientated tissue slice from an adult male pituitary. (B–E) Comparison of luminescence signal and luciferase protein localisation in single P1.5 pituitaries at low (B,C) and high (D,E) magnification. (B) Luminescence image of a whole P1.5 pituitary. (C) Immunofluorescence detection of luciferase protein in a P1.5 pituitary section (5 μm) generated from the pituitary imaged in B. Green shows luciferase protein, blue shows DAPI staining and yellow and/or red shows autofluorescence. (D) Luminescence image of the lateral part of a whole P1.5 pituitary gland. (E) Immunofluorescence detection of luciferase protein in a P1.5 pituitary section (5 μm) generated from the pituitary imaged in D. The colours are as described in C. (F,G) Luminescence images of P1.5 pituitaries in lateral and superior aspects, respectively. (H) Luminescence image of E19.5 pituitary. All luminescence images shown are from the start of organ culture. Immunofluorescence was performed following luminescence imaging and culture of pituitaries for 48 hours (24 hours in 10% FBS medium and 24 hours in 10% FBS medium supplemented with forskolin) (C, 5 μM forskolin; E, 1 μM forskolin). Scale bars: 200 μm.
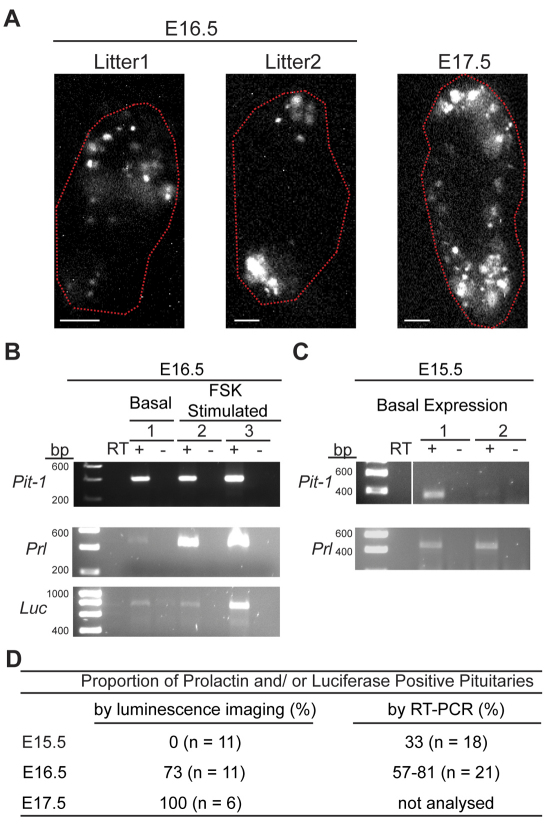
Fig. 2.
Onset of prolactin gene expression in developing pituitary. (A) Organisation of lactotroph cells in E16.5 and E17.5 pituitaries. The position of pituitary tissue, defined from brightfield images, is circled in red. Scale bars: 100 μm. Two E17.5 litters (12 pituitaries) and four E16.5 litters (28 pituitaries) were imaged. Images shown are from the start of organ culture. (B) RT-PCR detection of Pit-1, prolactin (Prl) and luciferase (Luc) from luminescent E16.5 pituitaries from litter 1 shown in A. Animal 1 was imaged for luminescence in basal culture medium (unsupplemented BSA medium) and then transferred into RNAlater solution within 6 hours. Animals 2 and 3 were imaged for 24 hours in rat serum medium followed by 1 μm forskolin (FSK) activation for 18 hours before transfer into RNAlater solution. Controls without reverse transcriptase (RT) are also shown. (C) RT-PCR for Pit-1 and Prl expression in undissected E15.5 pituitaries. Two pituitaries with prolactin expression are shown from 18 pituitaries analysed from two independent litters. (D) Table showing the proportion of pituitaries expressing prolactin and/or luciferase from E15.5 to E17.5 animals.
Transcription of prolactin in real-time in nascent lactotroph cells
Bioluminescence imaging of transgenic rat tissue provides a new technique, at single-cell resolution, by which to assess the onset of PRL gene transcription in the rat pituitary. To investigate this, whole hPRL-Luc49 pituitaries from various stages of embryonic development were imaged. E17.5 pituitaries, previously reported to be the earliest developmental stage at which prolactin is transcribed (Simmons et al., 1990), contained numerous luminescent cells at the periphery of the tissue (Fig. 2A). Imaging revealed fewer cells in E16.5 pituitaries (Fig. 2A), and no luminescent signal was detected from E15.5 pituitaries (data not shown). Nascent transcriptionally active lactotroph cells in E16.5 pituitaries were scattered throughout the pituitary tissue, and cells appeared to exist as isolated cells or small cell clusters (Fig. 2A). The presence of prolactin gene transcripts at E16.5 was confirmed by RT-PCR with primers against the endogenous rat Prl gene and the human-PRL-regulated luciferase reporter gene (Fig. 2B). Pit-1 transcripts, also detectable at this stage, were used as a positive control. Rat Prl transcripts were detectable at E15.5 by RT-PCR, but only at very low levels (Fig. 2C), requiring 400 ng of cDNA and 40 cycles of amplification for detection. The proportion of pituitaries that showed Prl and/or luciferase expression was also determined and increased with gestational age, becoming detectable in all pituitaries at E17.5 (Fig. 2D).
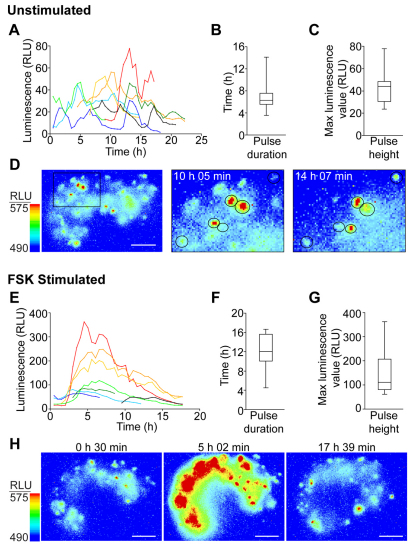
Bioluminescence imaging of transgenic E16.5 pituitaries was undertaken to determine the transcription pattern of an endocrine gene at the onset of transcription (Fig. 3). Photon counts were collected continuously for 24 hours, collating data over 30-minute intervals, from whole pituitaries cultured in a temperature-, CO2- and humidity-controlled imaging chamber. Intact pituitary tissue was used so that cells remained within their physiological context. To ensure that transcriptional function reflected the in vivo situation as faithfully as possible, we used culture medium containing 50% rat serum obtained from the dam; this was adapted from conditions previously shown to support whole embryo development in static culture systems (Jones et al., 2002). Bioluminescence imaging was performed for 24 hours in unstimulated conditions following which the pituitary tissue was treated with 1 μM forskolin, which caused an increase in luminescent signal, demonstrating that the cells remained viable in the culture conditions used. In unstimulated conditions, luminescence activity appeared highly pulsatile, with pulses varying in amplitude and duration (Fig. 3A–D). Forskolin stimulation of the tissue induced a transcriptional pulse from the majority of the detectable cells, with most cells responding at a similar time (Fig. 3E–H). The pulses had a variable duration and amplitude (Fig. 3F,G). Overall the data indicate that most transcriptionally active cells at E16.5 are competent to generate a pulsatile transcriptional response.
Fig. 3.
Single-cell transcription activity in nascent lactotrophs at E16.5. (A–D) Characterisation of transcription activity of lactotroph cells in E16.5 unstimulated pituitary tissue. (A) Luminescence activity in individual single cells from unstimulated E16.5 pituitary. (B) Box and whiskers plot of the distribution of transcription pulse duration in unstimulated E16.5 lactotroph cells. (C) Box and whiskers plot of the distribution of transcription pulse height in unstimulated E16.5 lactotroph cells. (D) Images of luminescent cells in E16.5 tissue. Left: whole E16.5 pituitary. The boxed area highlights cells that alter their transcription levels over time, as shown in the middle and right panels. (E–H) Characterisation of transcription activity of lactotroph cells in E16.5 pituitary tissue stimulated with forskolin (FSK) (1 μM). (E) Luminescence activity in individual single cells in forskolin-stimulated E16.5 pituitary tissue. (F) Box and whiskers plot of the distribution of transcription pulse duration in forskolin-stimulated E16.5 lactotroph cells. (G) Box and whiskers plot of the distribution of transcription pulse height in forskolin-stimulated E16.5 lactotroph cells. (H) Images showing changes in luminescence expression across the tissue over time following forskolin stimulation. Box and whisker plots depict the minimum and maximum values (lower and upper whiskers respectively), the first and third quartile (lower and upper limits of the box, respectively) and the median (solid line through the box). Scale bars: 100 μm. RLU, relative light units.
Transcription of prolactin in real-time in late embryonic and neonatal pituitary tissue
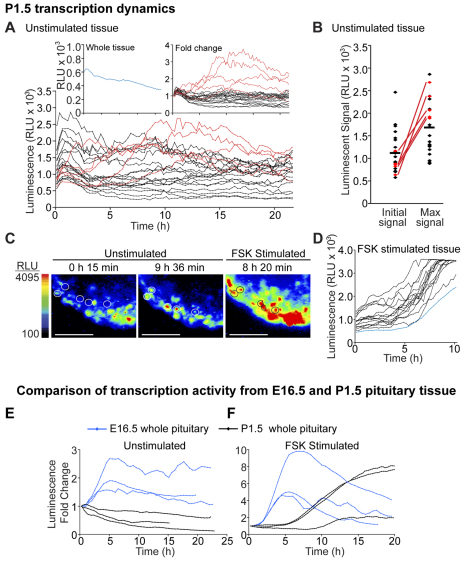
Bioluminescent imaging of transgenic E19.5 and P1.5 pituitaries was performed to assess whether PRL gene transcription patterns change during tissue development. Pituitaries were cultured in temperature-, CO2- and humidity-controlled imaging chambers in 50% rat serum medium, with photon counts collected continuously for 24 hours, collating data from either 15- or 30-minute intervals, depending on signal levels. The majority of cells in both E19.5 and P1.5 pituitary tissue showed a slight decline in luminescence levels over the observation period (Fig. 4A); luminescence from these cells was stable with no detectable pulses in activity. In P1.5 pituitary tissue 8% of cells (7 from 85 cells, range 6–20% in three independent experiments) showed transcriptional activation over the first 8 hours of culture, with increases in luminescent signal ranging from 1.4- to 3.1-fold (Fig. 4A). This transcriptional activation was rarely seen in cells from E19.5 pituitaries (1 from 83 cells, data not shown). Cells that showed the greatest transcriptional activation tended to have an initial low signal (Fig. 4B,C), which would be consistent with the release of these cells from dopaminergic inhibition in vivo (Hooghe-Peters et al., 1988; Nemeskeri et al., 1995). Alternatively, these cells could represent the most recently differentiated lactotroph cells, but this explanation is not entirely consistent with the increased number of these cells in neonatal pituitaries. Forskolin stimulation of the tissue induced a sustained increase in luminescence signal in most cells, with apparently similar dynamics (Fig. 4D). No differences in transcriptional patterns were seen between male- and female-derived pituitaries (data not shown).
Fig. 4.
Transcription activity changes during tissue development. (A–D) Characterisation of transcription activity in lactotroph cells in P1.5 pituitary tissue. (A) Graph comparing luminescence activity in 23 cells in P1.5 pituitary over 22 hours in 50% rat serum medium. Representative examples of single cells with stable luminescence activity (individual black lines) and cells that increase luminescence signal (individual red lines) are shown. Left inset: the overall luminescence signal from the whole pituitary tissue (blue). Right inset: data of single-cell activity depicted as fold change from the initial signal. (B) Graph depicting the level of luminescent signal emitted from each of the cells analysed at the start of imaging and their maximum signal emitted during imaging. Cells are depicted with the same colours in (A). The mean of the data is indicated by the horizontal line. (C) Images showing transcriptional activation of P1.5 cells (circled) in comparison with that in neighbouring cells in unstimulated conditions and during forskolin (FSK) (1 μM) stimulation. (D) Graph of luminescence signal from 15 single cells (individual black lines) and from the whole tissue (blue line) during forskolin (1 μM) stimulation. Data is representative of a total of 85 cells analysed from three independent pituitaries (two litters); data shown is from a male pituitary. (E,F) Comparison of transcription activity between P1.5 (black) and E16.5 (blue) whole pituitary tissue in unstimulated (E) and forskolin (1 μM)-stimulated (F) tissue (note: different scales are used in the graphs). Scale bars: 200 μm. RLU, relative light units.
The transcription patterns seen in nascent lactotrophs in E16.5 pituitary tissue were distinct from the transcription patterns observed in E19.5 and P1.5 pituitary tissue. This is reflected in the overall transcription pattern from the whole tissue, with E16.5 tissue showing an initial increase in luminescence signal and P1.5 tissue showing a slight gradual decrease in luminescence activity in unstimulated conditions (Fig. 4E). These patterns also differed from that of adult pituitary tissue (Harper et al., 2010). Differences could also be detected in the transcription patterns of the tissue (Fig. 4F) and individual cells (Fig. 3E; Fig. 4D) following forskolin stimulation. Overall this indicates that lactotroph cells alter their transcription pattern depending upon their developmental context.
Tissue architecture influences pulsatile prolactin transcription
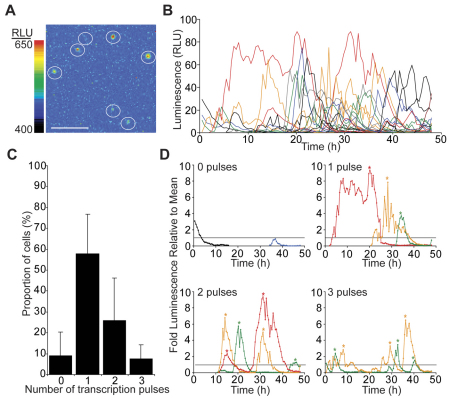
To assess whether PRL gene transcription is inherently stable in P1.5 cells or whether it is facilitated by tissue architecture or paracrine cell signalling, we enzymatically disaggregated individual pituitaries from hPRL-Luc49 animals, followed by luminescence imaging of the dispersed cell culture, maintained in 50% rat serum medium. Pulsatile luminescence activity was observed in the majority of individual cells (~90%), which showed large fluctuations in luminescence activity over several hours (Fig. 5A,B). Cells showed variable luminescence activity, undergoing between zero and three luminescence pulses in a 48-hour time period (Fig. 5C,D). Luminescence pulses were also highly variable with a large range in amplitude (1.2–8.4-fold above average signal) and duration (7–30 hours) (Fig. 5D). Similar patterns of pulsatile luminescence, and similar proportions of cells, were seen in cells cultured in 10% fetal bovine serum (FBS), albeit with higher baseline expression (supplementary material Fig. S1). This indicates that the induction of pulsatile transcription upon cellular isolation is not due to limited nutrients in the culture medium. Dispersed cell cultures generated from adult pituitaries have been imaged previously (Harper et al., 2010). Dispersed adult cells also show transcriptional pulses although with potentially different pulse dynamics (for example pulse amplitude). Overall, the induction of transcriptional pulse activity in dispersed cell cultures from P1.5 tissue indicates that tissue architecture or local cell signalling influences PRL transcriptional activity within developing pituitary tissue.
Fig. 5.
Isolated lactotroph cells, from enzymatically dispersed P1.5 pituitary tissue, show pulsatile luminescence activity. (A) Luminescence image showing the range of luciferase expression from enzymatically dispersed lactotroph cells. Cells are circled. Scale bar: 200 μm. (B) Graph showing luminescence activity of all cells from a single experiment. The luminescence of the whole field of view ranged from 1.24 to 1.59 relative light units (RLU). (C) Graph of the number of luminescence pulses that occurred in individual lactotroph cells over 48 hours from 57 cells analysed from four independent experiments (data are represented as means ± s.d.). (D) Graphs showing luminescence activity of representative cells with different numbers of luminescence pulses from a single experiment. The data were standardised to the mean luminescence level (set at 1, and shown by a horizontal line). Changes in signal below this level were not considered to constitute a transcriptional pulse. Changes in luminescence signal were also only considered as a transcriptional pulse if they doubled in signal intensity from the minimum signal value. The peak of each luminescence pulse is marked by an asterisk.
Discussion
Investigations into the dynamics of gene expression have, in the main, been limited to clonal cell lines. Recently, we reported the spatial and temporal characterisation of PRL gene expression within living adult pituitary tissue (Harper et al., 2010). Here, we extend these investigations to assess how tissue development impacts upon transcriptional activity. Using bioluminescence imaging of pituitary glands from transgenic hPRL-Luc49 Fischer-344 rats we assessed the transcriptional activity of the PRL gene in single living cells in fetal and neonatal tissue. Using this approach, we have shown that PRL transcription dynamics change during pituitary development, with several different phases of transcriptional activity. In newly differentiated lactotrophs, transcription rapidly turns on and off in individual cells, creating transient pulses of expression. Later in gestation and in early neonatal life, as PRL expression levels increase (Simmons et al., 1990), transcriptional activity appears to become more continuous, generating stable levels of expression.
Our data has important implications for the differentiation of lactotroph cells within the developing pituitary. The progression of lactotroph cells from a transient PRL-expressing stage early in development to a stable expressing population might reflect an important transition in cell differentiation. We propose that pulsatile gene expression early in tissue development might occur before lineage commitment, when numerous genes might exist in a poised state and can be transcribed at low levels. Commitment to a cell lineage leads to higher expression of the required genes and silencing of all other genes. This hypothesis is supported by observations that chromatin structure exists in a globally open and plastic conformation in embryonic stem cells, adopting a more closed structure upon cell differentiation (Mattout and Meshorer, 2010), and by the expression of genes from multiple lineages within single cells before lineage commitment (Hu et al., 1997; Efroni et al., 2008). Changes to chromatin structure have been shown to be important for the terminal differentiation of pituitary cells (Wang et al., 2007), thus transcription dynamics might change coincidently with chromatin structure during cell differentiation. Determining the factors that alter the transcription dynamics of the PRL gene early in development will be challenging owing to the difficulty of detecting lactotroph cells at this stage, the low number of lactotroph cells and the mixed cell populations that exist in the pituitary. We also detected a change in PRL gene transcription activity in early neonatal life, which we suspect is due to changes in key transcriptional regulatory mediators, such as oestrogen and dopamine, at this time (Slabaugh et al., 1982; Hooghe-Peters et al., 1988). Our data have further implications for the differentiation pathway of lactotroph cells because we are able to detect these cells at stages of development earlier than previously reported (Simmons et al., 1990). This indicates that lactotroph cells differentiate from Pit-1-positive precursor cells in a reduced timeframe. Lactotroph cells have been suggested to differentiate from somatotroph cells (Frawley, 1989), although the universal use of this pathway has been questioned (Luque et al., 2007). The short timeframe required for lactotroph differentiation argues against the frequent use of this indirect differentiation pathway.
The transient transcription pulses observed in nascent lactotrophs early in pituitary tissue development represent a simple transcriptional pattern. By contrast, lactotroph cells in adult pituitary display different complex patterns of transcriptional activity (Harper et al., 2010), which could be related to the functional heterogeneity of the cells. Nascent lactotroph cells are likely to be less functionally heterogeneous, consistent with the morphological similarity of the cells (Takahashi and Miyatake, 1991). Thus, the variability in the duration and amplitude of the transcription pulse in nascent lactotrophs, in unstimulated and stimulated conditions, might reflect the variable activity of the transcription machinery rather than major differences in the cell phenotype. Changes in cellular transcription activity, but not transcription pulse duration or pulse intensity, have been detected during Dictyostelium development (Chubb et al., 2006). In mammalian cell lines, characterisation of transcriptional ‘bursting’ has indicated important roles for the levels of transcription factors (Raj et al., 2006), promoter structure (Suter et al., 2011) and chromatin remodelling (Harper et al., 2011) in determining transcriptional activity, indicating that transcription pulses can be potentially altered and are not deterministic. Here, we report, for the first time, the quantitative assessment of transcriptional dynamics in developing fetal tissue and show that transcription patterns change during tissue development.
Tissue architecture might have an important influence on cellular transcription activity. We show that the stable transcription activity seen in neonatal pituitary lactotroph cells is lost upon enzymatic dispersal of the tissue. This has important implications for the study of transcription activity in isolated cells in vitro as this might differ substantially from the in vivo situation. Transcriptionally active lactotroph cells also show a specific topographical organisation within the pituitary, which is set up soon after lactotrophs first appear. This organisation of lactotroph cells might be important for lactotroph function, and again suggests that tissue structure has an important influence on lactotroph transcription activity. Comparison of cellular transcription profiles in E16.5 pituitary tissue indicated that some cells situated close together (within 35 μm) display similar patterns of activity, whereas most cells showed dissimilar transcription profiles (supplementary material Fig. S2). This could be owing to either paracrine cell signalling or to the functional similarity of the cells, possibly owing to recent derivation from the same progenitor cell. Functional cell networks have been detected in the pituitary gland and are established early in tissue development (Fauquier et al., 2001; Bonnefont et al., 2005), but whether such networks can coordinate transcription activity remains to be demonstrated. Lactotroph cells might exist as a functional cell network, although this has yet to be confirmed, but, in adult tissue, there is no evidence of correlated transcriptional activity between individual neighbouring cells (Harper et al., 2010).
In summary, we have characterised PRL gene transcription activity in the developing pituitary and have shown that nascent lactotroph cells progress from a pulsatile to a stable transcription phenotype. Analyses were performed on individual cells in intact tissue, and we show that tissue environment might have profound effects on transcriptional activity. Such single-cell analyses are crucial for determining the range and pattern of transcription activity within a tissue, and might provide insights into transcriptional mechanisms and the coordination of cellular activity to generate integrated tissue responses.
Materials and Methods
Animals
The generation and characterisation of Fischer 344 transgenic rats with the luciferase gene under the control of the human PRL locus (hPRL-Luc49) was described previously (Semprini et al., 2009). Animal studies were performed under UK Home Office license following review by the local ethics committee. Animals were housed in humidity- and temperature-controlled conditions (50±10% humidity; 20±1°C) in a 12-hour-light–12-hour-dark cycle. Animals were given free access to water and food (rat and mouse standard diet; SDS). Timed matings were set up with transgenic males and wild-type females with the detection of a vaginal plug the morning after mating considered as E0.5. Animals were killed at ~09:00 on the day of use by a schedule 1 method. Pituitaries were dissected from embryonic and neonatal rats and placed in either BSA medium [Dulbecco's modified Eagle's medium (DMEM) and F12, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM ultraglutamine, 0.25% (w/v) BSA, 1 mM luciferin (Biosynth)] or FBS medium [DMEM with 4.5 g/l glucose, 10% (v/v) FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM ultraglutamine, 1 mM luciferin]. Pituitaries placed in BSA medium were subsequently imaged in rat serum medium [DMEM and F12, 50% (v/v) dam serum, 100 U/ml penicillin, 100 μg/ml streptomycin, 2 mM ultraglutamine, 1 mM luciferin]. Rat serum used in culture was generated from trunk blood collected from the dam, following death, by exposure to a rising concentration of CO2 and cervical dislocation.
Embryonic and neonatal animals were sexed and genotyped by PCR. DNA was extracted from liver or tail tissue and sexed by PCRs against Sex-determining region Y (Sry), located on chromosome Y, and Testis specific X-linked gene (Tsx), located on chromosome X, and genotyped by PCRs against luciferase and renin. Primers used were: Sry forward, 5′-TGCAATGGGACAACAACCTA-3′ and Sry reverse, 5′-TAGTGGAACTGGTGCTGCTG-3′; Tsx forward, 5′-TTGGCCTCATGCATTACAAA-3′ and Tsx reverse, 5′-TCAACCTTCCAAAAGCAACA-3′; renin forward, 5′-CCTGGCAGATCACAATGAAGG-3′ and renin reverse 5′-GCATGATCAACTACAGGGAGC-3′; and luciferase forward, 5′-TTGCCAAGAGGTTCCATCTG-3′ and luciferase reverse 5′-GTCCAAACTCATCAATGTATC-3′.
Real-time bioluminescent imaging of pituitary glands
Whole embryonic or neonatal pituitaries were transferred into 35-mm dishes with glass coverslip bases (Iwaki) in BSA medium or FBS medium. Pituitaries were screened for luminescence using a Fluar 5× magnification 0.25 NA objective and captured using a photon-counting charge-coupled device camera (EBCCD; Hamamatsu Photonics). Pituitaries with luminescence activity to be imaged over time were transferred into a new 35-mm dish with either fresh FBS medium or rat serum medium. Imaging was performed using a Carl Zeiss Axiovert microscope in a dark room, maintained at 37°C in a Zeiss incubator with 5% CO2, 95% air and humidity. Luminescence images were captured using an Orca II CCD camera (Hamamatsu Photonics) or photon-counting charge-coupled device camera (EBCCD; Hamamatsu Photonics). Fluar 5× magnification 0.25 NA; 10× magnification 0.5 NA; or 20× magnification 0.75 NA objectives were used depending on pituitary size. Sequential luminescence images were taken with photons collected in 15–30-minute intervals depending upon the luminescent intensity of the pituitary. Brightfield images were taken before and after luminescence imaging. Pituitaries were activated with forskolin (1 μM), added directly to the dish, after 24 hours in basal culture medium. Images were taken and analysed using AQM Advance6 (Andor QI). The luminescence activity of whole pituitaries or individual cells was determined by subtracting the average instrument noise from the mean intensity of the selected region of interest.
Real-time bioluminescent imaging of pituitary primary cultures
P1.5 pituitaries with luminescent signal, detected by imaging, were disaggregated individually in HBSS supplemented with 0.1% (w/v) trypsin, 0.0045% (w/v) DNase I and 0.325% (w/v) BSA for 1.5 hours at 37°C. Cells were centrifuged at 250 g for 5 minutes and washed in HBSS containing 1% BSA. Cells were resuspended in rat serum medium or FBS medium and plated in cloning rings (4.7 mm diameter) in 35-mm coverslip based dishes (Iwaki). Cells were allowed to attach to the culture dish for 24 hours before the cloning ring was removed and additional medium added. Luciferin (1 mM) was added to the medium when the cells were put on the microscope for luminescence imaging, 38 to 40 hours after the cells were plated on culture dishes. Cells were imaged as described above using either a Fluar 10× magnification 0.5 NA or a 20× magnification 0.75 NA objective. Sequential luminescence images were taken with photons collected in 30-minute intervals for 48 to 72 hours. Luminescent signal intensity was determined in the same manner as for pituitary tissue.
Immunofluorescence
Pituitaries were fixed in 4% paraformaldehyde for 1 hour, embedded in wax and 5- μm sections cut and mounted onto poly-L-lysine slides (Thermofisher). Immunofluorescent staining was performed as described previously (Semprini et al. 2009). Mouse anti-luciferase (P. pyralis) antibody (Invitrogen/Zymed) was used at 1:20 and tissues were blocked in PBS supplemented with 20% (v/v) donkey serum and 5% (w/v) BSA. The luciferase antibody was detected with donkey anti-mouse-IgG antibody conjugated to Alexa Fluor 488 (1:200; Invitrogen) and sections were counterstained with DAPI. Slides were examined and images taken using a Nikon C1 confocal on an upright 90i microscope with 20× magnification 0.5 NA or 40× magnification 0.75 NA Plan Fluor objectives.
RT-PCR
Pituitaries were stored in RNAlater (Ambion) at −80°C until total RNA was extracted using the RNeasy kit (Qiagen). DNA was removed by DNase digestion using the RQ1 DNase I kit (Promega) followed by repurification of the RNA using the RNeasy kit and RNA cleanup protocol (Qiagen). Reverse transcription reactions (Roche) were performed in duplicate, with and without enzyme, on each RNA sample with oligo(dT). For dissected pituitaries, the whole RNA sample was used in the two reverse transcription reactions (20 ng–800 ng RNA per reaction) with bacteriophage MS2 RNA (200 ng; Roche) included in the reactions. For RNA extracted from pituitaries in situ, up to 4 μg of RNA was used per reaction. Primers used in PCRs were: Pit-1 forward 5′-AGGAACTCAGGCGGAAAAGT-3′ and Pit-1 reverse 5′-GAAGGTTTGCTGTGCTCTCC-3′; rat Prl forward 5′-CGGAAGTTCTTTTGAACCTGA-3′ and rat Prl reverse 5′-CTTAAGCATGTGCTGAAAGTTG-3′; luciferase primers used were those used for genotyping, detailed above.
Supplementary Material
Acknowledgements
We thank Ann Cookson and Catherine Savage for providing experimental assistance, and Anne Warhurst of the Manchester University Histology Facility for providing technical assistance. We also thank Peter March and Robert Fernandez of the Manchester University Bioimaging Facility for providing technical assistance with the Nikon C1 upright confocal microscope, purchased with grants from the BBSRC, Wellcome Trust and Manchester University Strategic Fund.
Footnotes
Funding
This work was supported by the Wellcome Trust [grant numbers 067252, 082794]; the Society for Endocrinology, UK; C.V.H. was funded by the Professor John Glover Memorial Postdoctoral Fellowship; S.S. was funded by a Wellcome Trust Intermediate Fellowship and a Research Excellence Award. The Developmental Biomedicine Research Group is supported by the Manchester Academic Health Sciences Centre (MAHSC) and the NIHR Manchester Biomedical Research Centre. The Centre for Cell Imaging has been supported through the BBSRC REI [grant number BBE0129651]; Hamamatsu Photonics and Carl Zeiss Limited provided technical support. Deposited in PMC for release after 6 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.088500/-/DC1
References
- Blake W. J., M, K. A., Cantor C. R., Collins J. J. (2003). Noise in eukaryotic gene expression. Nature 422, 633-637 [DOI] [PubMed] [Google Scholar]
- Bonnefont X., Lacampagne A., Sanchez-Hormigo A., Fino E., Creff A., Mathieu M. N., Smallwood S., Carmignac D., Fontanaud P., Travo P., et al. (2005). Revealing the large-scale network organization of growth hormone-secreting cells. Proc. Natl. Acad. Sci. U S A 102, 16880-16885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boockfor F. R., Frawley L. S. (1987). Functional variations among prolactin cells from different pituitary regions. Endocrinology 120, 874-879 [DOI] [PubMed] [Google Scholar]
- Camper S. A., Saunders T. L., Katz R. W., Reeves R. H. (1990). The Pit-1 transcription factor gene is a candidate for the murine Snell dwarf mutation. Genomics 8, 586-590 [DOI] [PubMed] [Google Scholar]
- Christian H. C., Chapman L. P., Morris J. F. (2007). Thyrotrophin-releasing hormone, vasoactive intestinal peptide, prolactin-releasing peptide and dopamine regulation of prolactin secretion by different lactotroph morphological subtypes in the rat. J. Neuroendocrinol 19, 605-613 [DOI] [PubMed] [Google Scholar]
- Elowitz M. B., Levine A. J., Siggia E. D., Swain P. S. (2002). Stochastic gene expression in a single cell. Science 297, 1183-1186 [DOI] [PubMed] [Google Scholar]
- Fauquier T., Guerineau N. C., McKinney R. A., Bauer K., Mollard P. (2001). Folliculostellate cell network: a route for long-distance communication in the anterior pituitary. Proc. Natl. Acad. Sci. U S A 98, 8891-8896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frawley L. S. (1989). Mammosomatotropes: current status and possible functions. Trends Endocrinol. Metab. 1, 31-34 [DOI] [PubMed] [Google Scholar]
- Harper C. V., Featherstone K., Semprini S., Friedrichsen S., McNeilly J., Paszek P., Spiller D. G., McNeilly A. S., Mullins J. J., Davis J. R., et al. (2010). Dynamic organisation of prolactin gene expression in living pituitary tissue. J. Cell Sci. 123, 424-430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper C. V., Finkenstadt B., Woodcock D. J., Friedrichsen S., Semprini S., Ashall L., Spiller D. G., Mullins J. J., Rand D. A., Davis J. R., et al. (2011). Dynamic analysis of stochastic transcription cycles. PLoS Biol 9 (4), e1000607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooghe-Peters E. L., Belayew A., Herregodts P., Velkeniers B., Smets G., Martial J. A., Vanhaelst L. (1988). Discrepancy between prolactin (PRL) messenger ribonucleic acid and PRL content in rat fetal pituitary cells: possible role of dopamine. Mol. Endocrinol. 2, 1163-1168 [DOI] [PubMed] [Google Scholar]
- Jones E. A., Crotty D., Kulesa P. M., Waters C. W., Baron M. H., Fraser S. E., Dickinson M. E. (2002). Dynamic in vivo imaging of postimplantation mammalian embryos using whole embryo culture. Genesis 34, 228-235 [DOI] [PubMed] [Google Scholar]
- Larson D. R., Singer R. H., Zenklusen D. (2009). A single molecule view of gene expression. Trends Cell Biol. 19, 630-637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Crenshaw E. B., 3rd, Rawson E. J., Simmons D. M., Swanson L. W., Rosenfeld M. G. (1990). Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene pit-1. Nature 347, 528-533 [DOI] [PubMed] [Google Scholar]
- Luque R. M., Amargo G., Ishii S., Lobe C., Franks R., Kiyokawa H., Kineman R. D. (2007). Reporter expression, induced by a growth hormone promoter-driven Cre recombinase (rGHp-Cre) transgene, questions the developmental relationship between somatotropes and lactotropes in the adult mouse pituitary gland. Endocrinology 148, 1946-1953 [DOI] [PubMed] [Google Scholar]
- McFerran D. W., Stirland J. A., Norris A. J., Khan R. A., Takasuka N., Seymour Z. C., Gill M. S., Robertson W. R., Loudon A. S., Davis J. R., et al. (2001). Persistent synchronized oscillations in prolactin gene promoter activity in living pituitary cells. Endocrinology 142, 3255-3260 [DOI] [PubMed] [Google Scholar]
- Nemeskeri A., Acs Z., Toth B. E. (1995). Prolactin-synthesizing and prolactin-releasing activity of fetal and early postnatal rat pituitaries: in vivo and in vitro studies using RIA, reverse hemolytic plaque assay and immunocytochemistry. Neuroendocrinology 61, 687-694 [DOI] [PubMed] [Google Scholar]
- Newlands S., Levitt L. K., Robinson C. S., Karpf A. B., Hodgson V. R., Wade R. P., Hardeman E. C. (1998). Transcription occurs in pulses in muscle fibers. Genes Dev 12, 2748-2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare A., Lemons D., Kosman D., Beaver W., Freund Y., McGinnis W. (2009). Visualization of individual Scr mRNAs during Drosophila embryogenesis yields evidence for transcriptional bursting. Curr. Biol. 19, 2037-2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raser J. M., O'Shea E. K. (2004). Control of stochasticity in eukaryotic gene expression. Science 304, 1811-1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semprini S., Friedrichsen S., Harper C. V., McNeilly J. R., Adamson A. D., Spiller D. G., Kotelevtseva N., Brooker G., Brownstein D. G., McNeilly A. S., et al. (2009). Real-time visualization of human prolactin alternate promoter usage in vivo using a double-transgenic rat model. Mol. Endocrinol. 23, 529-538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons D. M., Voss J. W., Ingraham H. A., Holloway J. M., Broide R. S., Rosenfeld M. G., Swanson L. W. (1990). Pituitary cell phenotypes involve cell-specific Pit-1 mRNA translation and synergistic interactions with other classes of transcription factors. Genes Dev. 4, 695-711 [DOI] [PubMed] [Google Scholar]
- Stirland J. A., Seymour Z. C., Windeatt S., Norris A. J., Stanley P., Castro M. G., Loudon A. S., White M. R., Davis J. R. (2003). Real-time imaging of gene promoter activity using an adenoviral reporter construct demonstrates transcriptional dynamics in normal anterior pituitary cells. J. Endocrinol 178, 61-69 [DOI] [PubMed] [Google Scholar]
- Suter D. M., Molina N., Gatfield D., Schneider K., Schibler U., Naef F. (2011). Mammalian genes are transcribed with widely different bursting kinetics. Science 332 (6028): 472-474 [DOI] [PubMed] [Google Scholar]
- Takahashi S., Miyatake M. (1991). Immuno-electron microscopical study of prolactin cells in the rat. Postnatal development and effects of estrogen and bromocriptine. Zool Sci. 8, 549-559 [Google Scholar]
- Takasuka N., White M. R., Wood C. D., Robertson W. R., Davis J. R. (1998). Dynamic changes in prolactin promoter activation in individual living lactotrophic cells. Endocrinology 139, 1361-1368 [DOI] [PubMed] [Google Scholar]
- Wijgerde M., Grosveld F., Fraser P. (1995). Transcription complex stability and chromatin dynamics in vivo. Nature 377, 209-213 [DOI] [PubMed] [Google Scholar]
- Zhu X., Gleiberman A. S., Rosenfeld M. G. (2007). Molecular physiology of pituitary development: signaling and transcriptional networks. Physiol. Rev. 87, 933-963 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.