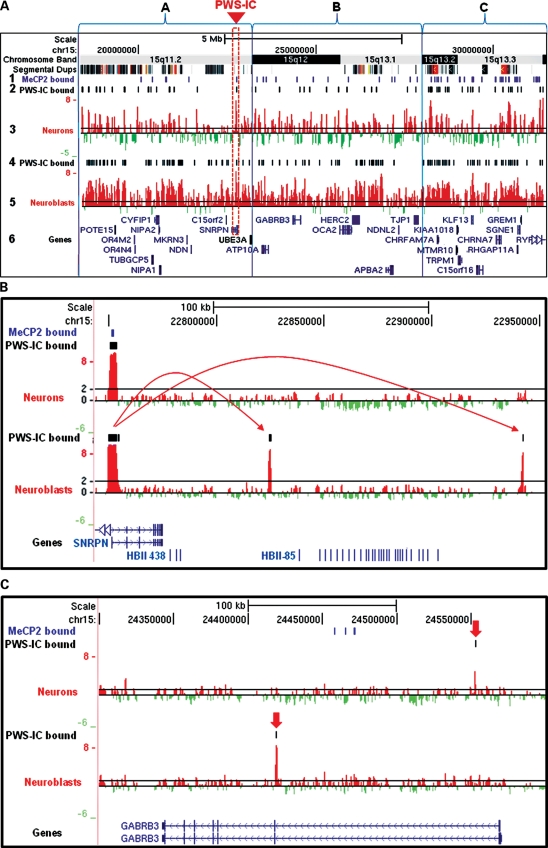
Figure 1.
Dynamic 15q11.2–13.3 chromatin decondensation during neuronal maturation. (A) 4C analysis of 15q11.2–13.3 chromatin structure in developing neurons shows developmentally regulated chromatin decondensation in maturing neurons. 15q11.2–13.3 loci that contact the PWS-IC (red box) are shown as a series of log 2 signal peaks from representative hybridization of 4C libraries isolated from developing SH-SY5Y neurons (row 3) and untreated SH-SY5Y neuroblasts (row 5). 4C peaks corresponding to sites of PWS-IC interaction were determined using a bioinformatic analysis of three independent replicate experiments for both PMA-treated neurons and untreated neuroblasts and are shown as a series of black bars above the corresponding peaks (rows 2 and 4). MeCP2-binding sites determined from previous studies are shown as a series of purple bars in row 1. 15q11.2–13.3 was subdivided into regions A–C based on Giemsa banding patterns in metaphase chromosomes (top row). Log 2 signal ratios are shown as red and green histograms in the UCSC Genome Browser with known genes and transcripts shown in blue font (row 6). A black line at a log 2 signal ratio of 1 is included to enable comparison of peaks in rows 3 and 5. Segmental duplications are shown as a series of colored bars below chromosome bands. (B). A high-resolution example of PWS-IC interactions flanking the HBII-85 snoRNA cluster in neuroblasts (bottom histogram) that are lost during neuronal differentiation (top histogram). Arrows show PWS-IC-binding interactions. (C). PWS-IC interactions shown as black lines above the log 2 signal peaks with the autism candidate gene GABRB3 are also altered by neuronal differentiation. PWS-IC interactions are indicated by red arrows.