Abstract
Although fungi contribute significantly to the microbial biomass in terrestrial ecosystems, little is known about their contribution to biogeochemical nitrogen cycles. Agricultural soils usually contain comparably high amounts of inorganic nitrogen, mainly in the form of nitrate. Many studies focused on bacterial and archaeal turnover of nitrate by nitrification, denitrification and assimilation, whereas the fungal role remained largely neglected. To enable research on the fungal contribution to the biogeochemical nitrogen cycle tools for monitoring the presence and expression of fungal assimilatory nitrate reductase genes were developed. To the ∼100 currently available fungal full-length gene sequences, another 109 partial sequences were added by amplification from individual culture isolates, representing all major orders occurring in agricultural soils. The extended database led to the discovery of new horizontal gene transfer events within the fungal kingdom. The newly developed PCR primers were used to study gene pools and gene expression of fungal nitrate reductases in agricultural soils. The availability of the extended database allowed affiliation of many sequences to known species, genera or families. Energy supply by a carbon source seems to be the major regulator of nitrate reductase gene expression for fungi in agricultural soils, which is in good agreement with the high energy demand of complete reduction of nitrate to ammonium.
Keywords: agriculture, fungi, gene expression, nitrate assimilation, nitrate reductase, soil
Introduction
Agricultural soils normally encounter regular nitrate (NO3−) flushes, either directly from nitrate fertilization or from ammonium (NH4+) or urea, which are rapidly converted to NO3− by nitrification. NO3− can subsequently be lost from agricultural systems via leaching to the groundwater or via denitrification to nitrous oxide (N2O) or dinitrogen (N2) to the atmosphere (Zumft, 1997). Both processes cause environmental problems and, moreover, consumption of NO3−-contaminated drinking water poses health risks to humans and animals and N2O is a potent greenhouse gas (for a recent summary on biogeochemical nitrogen cycles and their impact on the environment, see Galloway et al., 2008).
Both plants and phylogenetically diverse microorganisms have high NO3− assimilation capacities (Glass et al., 2002; Stitt et al., 2002; Inselsbacher et al., 2010). Incorporation of nitrogen from NO3− into microbial biomass leads to immobilization and therefore reduces losses through leaching or denitrification. The ability to assimilate NO3− is widespread among microbes. In the fungal kingdom, NO3− utilization is restricted to the Dikarya, that is, the Ascomycota and the Basidiomycota (Slot et al., 2007; Slot and Hibbett, 2007) and certain members from the Mucorales (Sarbhoy, 1965). NO3− assimilation is however not ubiquitous in the Dikarya with many nonutilizing species found especially among the yeasts (Lodder, 1970).
In many fungi, NO3− assimilation genes are clustered and horizontal gene transfer (HGT) from an oomycete to an ancestor of the Dikarya has been proposed (Slot et al., 2007; Slot and Hibbett, 2007). The presence of a nitrate reductase gene in the genome of Mucor circinelloides could, however, be indicative of an earlier acquisition of the NO3− assimilation cluster by fungi. Within the Dikarya, a secondary HGT event was proposed from a basidiomycetous donor to the ascomycetous genus Trichoderma (Slot and Hibbett, 2007).
Regulation of fungal NO3− assimilation has been extensively studied (Marzluf, 1997; Siverio, 2002) and is today best understood in the model ascomycete Aspergillus nidulans (Hynes, 1973; Cove, 1979; Punt et al., 1995; Muro-Pastor et al., 1999, 2004; Narendja et al., 2002; Berger et al., 2006, 2008; Bernreiter et al., 2007; Schinko et al., 2010). In general, NO3− uptake and assimilation in fungi is repressed in the presence of preferred nitrogen sources (Wong et al., 2008), but differences occur at the level of repression. Although NH4+ or glutamine completely repress uptake and utilization of any alternative nitrogen source in the model ascomycetes Neurospora crassa (Premakumar et al., 1979) and A. nidulans (Arst and Cove, 1973), a more relaxed mode of action was found in several Fusarium species (Celar, 2003) and in the yeast Candida nitratophila (Ali and Hipkin, 1986). Similar variations on the same theme are seen at the pathway-specific level: although the presence of NO3− (or nitrite) is obligatory for full induction of uptake and assimilation in N. crassa and A. nidulans (Hawker et al., 1992; Marzluf, 1997), this is not the case in the ectomycorrhizal basidiomycetes Hebeloma cylindrosporum (Jargeat et al., 2000, 2003) and Laccaria bicolor (Kemppainen et al., 2010) and the yeasts C. nitratophila and Sporobolomyces roseus (Ali and Hipkin, 1985).
In good agreement with studies on model organisms under laboratory conditions, assimilatory nitrate reductase activity in aerated soil slurries amended with glucose was found to be strongly inhibited by NH4+ and glutamine (McCarty and Bremner, 1992a, 1992b). On the other hand, high NO3− immobilization rates were found in grassland (Davidson et al., 1990) and arable (Inselsbacher et al., 2010) soils even in the presence of NH4+. Whereas the studies by McCarty and Bremner, (1992a, 1992b) employed measurement of NO3− removal in aerated soil slurries amended with glucose and NO3−, the study by Inselsbacher et al. (2010) measured incorporation of nitrogen-15 into microbial biomass in a soil microcosm system (Inselsbacher et al., 2009). Under these experimental conditions, microsites might occur where the repressive effect of NH4+ on NO3− assimilation is alleviated because of contrasting soil mobility of the two inorganic nitrogen forms (Davidson et al., 1990; Inselsbacher et al., 2010).
To characterize assimilatory nitrate reductases in environmental samples, tools and data are available for bacterial (Allen et al., 2001; He et al., 2007), archaeal (Alcántara-Hernández et al., 2009) and basidiomycetous (Nygren et al., 2008) genes. In contrast, the analogous tools and databases have not yet been available for ascomycetes, which constitute the dominant fungal group in many agricultural and grassland soils (Klaubauf et al., 2010). In this study we developed degenerate primers that allowed the amplification of nitrate reductase encoding genes (euknr) from fungi. The deduced protein sequences are consequently termed EukNR. This naming should avoid confusion with the bacterial nomenclature, where nar is used for the respiratory, nap for the periplasmic and nas for the assimilatory nitrate reductases (Richardson and Watmough, 1999). EukNR is structurally not related to bacterial nitrate reductases but rather to pro- and eukaryotic sulphite oxidases (Campbell, 2001). The main focus in primer design has been put on Ascomycota, and within the Ascomycota on the subphylum Pezizomycotina, which constitute the main group in agricultural soils (Klaubauf et al., 2010). The newly developed primers have been used on individual culture isolates to improve the reference database of euknr genes. In addition, total DNA isolated from agricultural soils has also been used as template in an attempt to gain a better insight into the fungal community able to utilize NO3−. Finally, gene expression of fungal euknr has been studied in a selected agricultural soil, revealing the most prominent members of the soil fungal community expressing nitrate reductase mRNA.
Materials and methods
Fungal strains and growth conditions
A complete list of fungal strains used in this study is given in Supplementary Table 1. All cultures were maintained at room temperature on malt extract agar (Merck, Darmstadt, Germany) except for Bionectria coronata CBS 696.93 and Bionectria grammicospora CBS 209.93, which were maintained on potato dextrose agar (Roth, Karlsruhe, Germany).
DNA extraction, PCR and sequencing
DNA extraction from axenically growing culture isolates was performed with the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. DNA from soils Maissau (soil M) and Purkersdorf (soil P) was taken from Klaubauf et al. (2010). DNA from experiments with soil from Niederschleinz (soil N) was eluted after RNA extraction (see below) with the RNA PowerSoil DNA Elution Accessory Kit (MO BIO, Carlsbad, CA, USA). For field site and soil sampling description, see Inselsbacher et al. (2009).
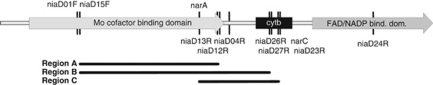
Primers for amplification of fungal euknr were designed based on multiple alignments of available full-size EukNR sequences. Primer details are given in Table 1 and Supplementary Table 2. Positions of primers in the euknr gene are shown in Figure 1. The routine amplification protocol employed a first PCR step with primer pair niaD01F/niaD04R and a nested PCR with primer pairs niaD15F/niaD12R (Ascomycota) or niaD15F/niaD13R (Basidiomycota), although from pure culture isolates single-round amplification with primer pairs niaD15F/niaD12R or niaD15F/niaD13R usually worked well. From Bionectria ochroleuca and Myxotrichaceae, longer PCR products were generated with more specific primers to improve phylogenetic placement. Primer variants with T7- or M13fwd-sequences attached to the 5′-end were used for direct sequencing of PCR products, as sequencing with highly degenerate primers often resulted in poor-quality reads. The PCR mixtures were as follows: 1 × GoTaq Green Master Mix (Promega, Mannheim, Germany), 2 μ of each primer, 0.5 mg ml–1 bovine serum albumin, 1.25 m additional MgCl2 and template. The cycling conditions were as follows: initial denaturation at 95 °C for 2 min and 30 s followed by 35 cycles of 94 °C for 20 s, 52 °C for 20 s and 72 °C for 1 min followed by a final extension at 72 °C for 5 s.
Table 1. Primers for routine amplification of fungal euknr from pure culture isolates and environmental samples.
| Primer | Sequence | Expected PCR product size |
|---|---|---|
| niaD01F | 5′-GTNTGYGCNGGNAA-3′ | |
| niaD04R | 5′-GTNGGRTGYTCRAA-3′ | niaD01F+niaD04R: 1 kb |
| niaD15F | 5′-GGNAAYMGNMGNAARGARCARAA-3′ | |
| niaD12R | 5′-AACCANGGRTTRTTCATCATNCC-3′ | niaD15R+niaD12R: 0.7–1 kb |
| niaD13R | 5′-GGTRGCGTTCCAGTACATRTC-3′ | niaD15R+niaD13R: 0.8–1 kb |
Figure 1.
Position of primers for amplification of euknr genes. Positions of primers for amplification of fungal euknr genes are shown on a schematic map of the EukNR domain structure. Forward primers are indicated above and reverse primers below the map; niaD primers are from this study, and nar primers from Nygren et al. (2008) are shown for comparison. For primer details, see Table 1 and Supplementary Table 2. Regions used for phylogenetic studies are indicated at the bottom. Region A was used for construction of the backbone tree and for phylogenetic placement of soil genomic and cDNA clones. Region B was used to obtain improved phylogenetic resolution of horizontal gene transfer events in the Trichoderma/Bionectria-clade and in the Myxotrichaceae (see text and Figure 2). Region C was used by Nygren et al. (2008) in their study of EukNR from ectomycorrhizal basidiomycetes.
Sequencing was done by AGOWA GmbH (Berlin, Germany). Accession numbers are HQ115646–HQ115737 for rRNA gene cluster sequences for identification of pure culture isolates, HQ234766–HQ234874 for partial euknr from pure culture isolates (see Supplementary Table 1), HQ243445–HQ243656 for partial euknr amplified from soil genomic DNA and HQ288852–HQ288891 for partial euknr amplified from soil complementary DNA (cDNA).
Microcosm experiment
The experimental system for analysing soil fungal RNA expression consisted of microcosms filled with agricultural soil N as described in Inselsbacher et al. (2009, 2010). Soil was amended with 1% (w/w on a fresh weight basis) of a cellulose/starch/xylan mixture before setup of the experiment. Treatment A received no additional organic fertilizer, treatment B received 0.05% (w/w on a fresh weight basis) Biovin (Intertrest, Guntramsdorf, Austria) and treatment C received 0.05% autoclaved Biovin. Biovin is an organic fertilizer produced from humified grapevine pomace. Each treatment was replicated ten times (five replicates for each harvest time point). Microcosms were equilibrated to 62% water-filled pore space, fertilized twice with potassium nitrate (KNO3) and planted with winter barley as described in Inselsbacher et al. (2009, 2010). Sampling time points for soil RNA isolation and determination of soil nitrogen pools corresponded to days 1 and 6 in Inselsbacher et al. (2010), that is, 1 and 6 days after planting the pregerminated seedlings and the second fertilization. Only microcosms with fully developed winter barley seedlings were used for further analyses. At the first sampling day, four A, five B and four C treatments and at the sixth day, four A, four B and five C treatments had been fully developed and were used for further analyses. Soil chemical analyses were done as described in Inselsbacher et al. (2010).
RNA isolation and cDNA synthesis
At harvest the whole content of one microcosm was transferred to a petri-dish, plant shoots were removed and plant roots were carefully separated from the soil. Soil was mixed and an aliquot (2 g fresh weight) was used for RNA isolation with the PowerSoil Total RNA Isolation Kit (MO BIO) according to the manufacturer's instructions with a minor modification: the organic phase and the interphase were re-extracted by addition of 2.5 ml Bead Solution, 0.25 ml SR1 and 0.8 ml SR2, followed by thorough vortexing and centrifugation. The aqueous layers from the first and second extraction were combined. Amounts of buffers were doubled for the subsequent steps until loading onto the RNA capture columns. Integrity of RNA was checked on a conventional 1.5% agarose gel, and the concentration determined spectrophotometrically with the Quant-iT RNA BR Kit (Invitrogen, Paisley, UK). From each replicate, 8 μl of total RNA (∼0.1 μg) was treated with 1 U DNase I (Invitrogen) in a volume of 10 μl for 15 min at room temperature. Reactions were terminated by addition of 1 μl 25 m EDTA and heating to 65 °C for 10 min. DNase-I-treated RNA was directly subjected to reverse transcription with random hexamer primers with RevertAid H− First Strand cDNA Synthesis Kit (Fermentas, St Leon-Rot, Germany) in a volume of 20 μl according to the manufacturer's instructions. Single-stranded cDNA was diluted to 100 μl and directly used for PCR with euknr primers as described above. Control reactions included PCR with untreated RNA and RNA after DNase I treatment as templates and PCR with primers EF1α983F/EF1α1567R for amplification of fungal translation elongation factor 1α (for primer details, see http://aftol1.biology.duke.edu /pub/primers/viewPrimers). No PCR products could be obtained from RNA after DNase I treatment but before reverse transcription.
At sampling time point day 1, nested PCR on cDNA with primers designed for ascomycetous euknr gave products of the expected size in two, four and three replicates from treatments A, B and C, respectively. PCR products from day 1 were pooled per treatment (A, B and C) before cloning. As from day 6, euknr could only be detected with a weak signal in 1 out of 13 RNA samples, this time point was omitted from further analyses.
Cloning
PCR products were cloned in plasmid pTZ57R/T (Fermentas) according to the manufacturer's instructions. Cloning of euknr-PCR products from pure culture isolates was only necessary in one case (strain Phialophora mustea KSS12_F02), which showed overlapping signals upon direct sequencing of PCR products. In all, 12 randomly selected clones were picked and sequenced from both ends with vector primers. For Cadophora finlandica, a genomic clone containing a nearly full-length euknr including the promoter region was isolated from an existing library (Gorfer et al., 2009) by PCR amplification with euknr and plasmid primers (Acc. Nr. HQ234780).
For analysis of fungal nitrate reductase genes in agricultural soils, euknr was amplified from genomic DNA from soils M and P with primer pairs designed for ascomycetous and basidiomycetous euknr, respectively. PCRs were run in triplicate and pooled before cloning. From each of the resulting four libraries, 96 clones were randomly picked. For soil N from the microcosm experiment, only the primer pair designed for the ascomycetous euknr was used. From each of the resulting three libraries, 72 clones were randomly picked. For analysis of euknr cDNA pools in soil N, only the primer pair designed for the ascomycetous euknr was used. Amplification was done separately on the replicates from harvest day 1 and pooled per treatment before cloning. From the pools A, B and C, 85, 46 and 39 clones were picked, respectively.
Clones were first screened by restriction fragment length polymorphism with the restriction enzyme BsuRI (Fermentas isoschizomere of HaeIII) and up to five randomly selected clones from each restriction fragment length polymorphism pattern were sequenced by vector primers from both ends.
Sequence analysis
Forward and reverse sequence reads were assembled using Vector NTI Advance 10 and 11 (Invitrogen). Introns were detected based on conserved intron positions, conserved boundary sequences and deduced amino-acid sequence after in silico splicing of the introns. Edited nucleotide sequences were translated to amino-acid sequences by Vector NTI. Multiple alignments of protein sequences (MUSCLE; Edgar, 2004) and construction of phylogenetic trees were done with molecular evolutionary genetics analysis version 4 (MEGA4; Tamura et al., 2007) and MEGA5 (Tamura et al., 2011). Maximum likelihood trees were calculated with the WAG+G model (Whelan and Goldman, 2001) and 500 bootstrap replicates.
Chimaeric reads from genomic and cDNA libraries were detected by Bellerophon (Huber et al., 2004) and removed before further analyses. For affiliation of soil genomic and cDNA clones, sequences were clustered with MOTHUR (Schloss et al., 2009) at 96% identity (furthest neighbour) at the protein level. At this cutoff, separation of most species was possible, although some species (for example, in the genus Tetracladium) still clustered together whereas, for example, Fusarium oxysporum is divided into two subgroups. With these settings, clones with identical BsuRI restriction patterns consistently clustered together.
Phylogenetic hypothesis testing
Four replicates each of constrained and unconstrained trees were calculated with RaxML 7.0.3 (Stamatakis, 2006) using the PROTMIXWAGF setting, an implementation of the WAG+G model of amino-acid sequence evolution (Whelan and Goldman, 2001), as selected with MultiPhyl v1.0.6 (Keane et al., 2007). Single-site likelihoods were calculated in RaxML using the PROTGAMMAWAGF setting and imported into CONSEL 0.1i (Shimodaira and Hasegawa, 2001) to compare the tree topologies by a series of statistical tests, including the approximately unbiased test of phylogenetic tree selection (Shimodaira, 2002).
Statistical analyses
Differences in soil nitrogen pools between soils were tested by one-way analysis of variance. Cluster analysis (nearest neighbour method with squared Euclidian distance metric) of euknr communities in soils M and P amplified with different primer pairs was performed to investigate the similarity between and within soil samples. All statistical analyses were run with StatGraphics version 5 (StatPoint Technologies, Inc., Warrenton, VA, USA).
Results
The phylogenetic tree of fungal EukNR sequences
The fungal EukNR tree is based on published full-length or nearly full-length sequences, mainly from fungal genome sequencing projects, but also from early studies on structure and function of EukNR. An overview of these sequences is given in Supplementary Table 3. The full-length data set consists of three Phytophthora sequences, which represent the putative oomycete donor of horizontally transferred fungal EukNR (Slot and Hibbett, 2007), 1 sequence from Mucoromycotina, 9 sequences from Basidiomycota and 72 sequences from Ascomycota. Additionally, one sequence from an Oryza sativa EST library (AK110249; Kikuchi et al., 2003) was included. This sequence obviously encodes a EukNR from a fungus related to Ustilago maydis, which had apparently infected the analysed rice cultivar. No corresponding sequence was found in the rice genome, excluding the possibility that the sequence was obtained by HGT. Phylogenetic analysis of the full-length data set provides evidence that euknr was transferred from an oomycete donor to a common ancestor of Mucorales and Dikarya (95% bootstrap support for placing M. circinelloides EukNR sister to dikaryan EukNR; see Supplementary Figure 1). The rare but evenly spread distribution of nitrate assimilation in the Mucorales (cf Domsch et al., 1993) suggests frequent losses of euknr from mucoralean genomes, as for example in Rhizopus oryzae (Ma et al., 2009). More sequence data from the basal fungal lineages are needed for a reliable reconstruction of the gene gains and losses that finally led to today's distribution of euknr in the fungal kingdom.
As many fungal species commonly found in agricultural soils have no close relatives with completely sequenced genomes, additional euknr sequences were generated from pure culture isolates. To this end, 115 ascomycetes and 3 basidiomycetes were analysed for the presence of euknr by PCR. From 104 ascomycetes and 3 basidiomycetes, euknr could be amplified. All PCR products showed highest similarity to fungal nitrate reductases, and not to any other proteins also containing the Molybdenum cofactor like sulphite oxidases. In only two cases, gene duplication of euknr was found: for B. ochroleuca Rd0801 (see below) and Phialophora mustea KSS12_F02. EukNRs from P. mustea KSS12_F02 have 78.8% sequence identity and cluster together in a sub-branch of the Hypocreales (see Supplementary Figure 2). In both copies, essential residues for binding of Molybdenum cofactor are conserved. This pattern is indicative for a gene duplication in a recent ancestor of P. mustea KSS12_F02.
Overall, there is good congruence of the fungal EukNR tree with widely accepted phylogenies (Hibbett et al., 2007). In the EukNR tree based on the full-length sequences, all orders appear monophyletic, although the Hypocreales (excluding Trichoderma, see Slot and Hibbett, 2007) and Onygenales lack bootstrap support (see Supplementary Figure 1). The EukNR tree based on the partial Molybdenum cofactor-binding domain (region A in Figure 1) provides increased phylogenetic coverage because of inclusion of additional taxa, but shows decreased resolution at certain nodes (Supplementary Figure 2).
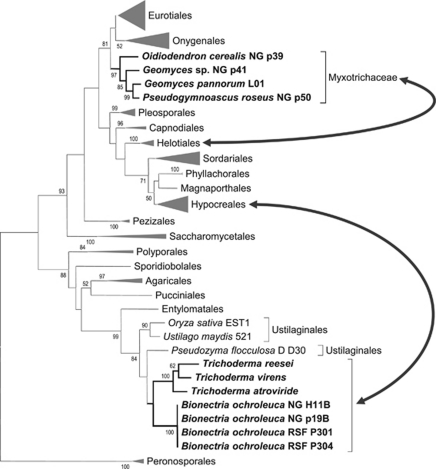
In two cases, indications of HGT within the Dikarya could be found. Although acquisition of a nitrate assimilation gene cluster in the genus Trichoderma from a basidiomycete related to Ustilago via HGT was already previously described (Slot and Hibbett, 2007), the origin and destination of this cluster could now be narrowed down. According to our phylogenetic data, the donor belonged to the Ustilaginaceae and the acceptor was a common ancestor of Trichoderma/Hypocrea and Bionectria (Figure 2). As one out of seven analysed B. ochroleuca strains contained two euknr genes—one ascomycetous and one basidiomycetous—it seems likely that HGT occurred before loss of the autochthonous euknr. Presence of an ascomycetous euknr in the remaining B. ochroleuca strains cannot be excluded, but in none of the completely sequenced Trichoderma genomes an ascomycetous euknr could be found.
Figure 2.
Horizontal gene transfer of euknr. The EukNR phylogenetic tree provides evidence for horizontal gene transfer of euknr from a basidiomycete related to the Ustilaginaceae to a common ancestor of the ascomycetes Trichoderma and Bionectria (Hypocreales; Slot and Hibbett, 2007 and this study) and from an ascomycete related to the Eurotiales and Onygenales to an ancestor of the Myxotrichaceae, which are closely related to the Helotiales (this study). Tree branches affected by gene transfer events are highlighted in bold. The arrows indicate different positioning of the involved branches between accepted phylogenies and the EukNR tree (shown here). The phylogenetic tree is based on the partial sequence of the Molybdenum cofactor (MoCo)-binding domain and the partial cytochrome b5 domain (region B in Figure 1) of available full-length or nearly full-length EukNR sequences and partial sequences from Trichoderma, Bionectria and the Myxotrichaceae. Where appropriate, branches were collapsed at the ordinal level (grey triangles) to reduce complexity of the image. Bootstrap support for branches above 50% is shown.
Another HGT event seems to have occurred within the Ascomycota. EukNR from the Myxotrichaceae groups with the Eurotiales and Onygenales. Formerly the genera Geomyces, Pseudogymnoascus and Oidiodendron were placed in the Onygenales (Currah, 1985), although phylogenetic studies recently moved this family close to the Helotiales within the Leotiomycetes (Tsuneda and Currah, 2004; Wang et al., 2006a, 2006b). There is, however, excellent bootstrap support for placement of EukNR from G. pannorum L01, Geomyces sp. NG_p41, P. roseus NG_p50 and O. cerealis NG_p39 sister to the Eurotiales and Onygenales, and additional support comes from intron numbers and positions in the amplified euknr fragment: although all analysed euknr genes in the subclass Eurotiomycetidae and in the family Myxotrichaceae show three introns at conserved positions within the amplified fragment, no intron is found in the corresponding region in the Helotiales (see Supplementary Figure 3). Constrained trees, where EukNR from Myxotrichaceae was forced to group with the Helotiales, were rejected by all topological tests applied, including the conservative weighted Shimodaira–Hasegawa test (Shimodaira and Hasegawa, 2001).
Fungal euknr genes in agricultural soils
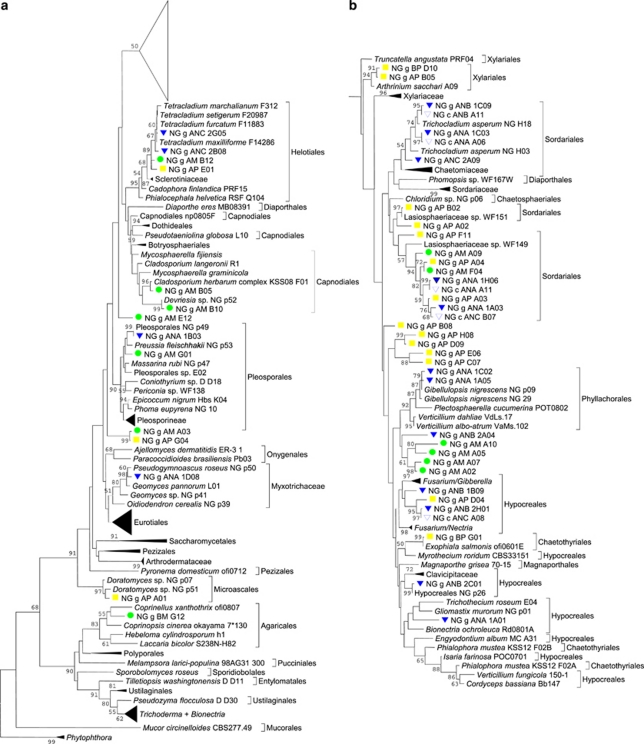
Agricultural soil fungal communities with the potential to assimilate NO3− were studied by direct amplification of fungal euknr genes from total soil DNA. The following soils from Lower Austria were analysed: Maissau (soil M), Purkersdorf (soil P) and Niederschleinz (soil N). For details on treatment of soil N, see Materials and methods. Soils M and P were analysed in parallel with primers designed for ascomycetous and basidiomycetous euknr, respectively. Surprisingly, both primer pairs amplified mainly ascomycetous euknr genes from soils M and P. Only one basidiomycetous euknr gene affiliated to Coprinellus was detected in soil M. Libraries amplified from the same soil had a significantly lower Euclidian distance (that is, were significantly more similar) than libraries amplified from the two different soils. Therefore, data were merged to two soil-specific data sets. Based on results from soils M and P, only the primer pair designed for the ascomycetous euknr was applied to soil N. Although some sequences from the genomic libraries could be clearly assigned to a distinct species or genus, the majority of sequences could only be assigned to higher taxonomic ranks (see Figure 3 and Table 2). From sequences that showed clear affiliation at the ordinal level, the majority belonged to the Sordariales, Hypocreales and Helotiales. In soil N, however, a large number of clones (39%) belonged to the Peronosporales, and one sequence clustered with M. circinelloides. These results indicate that the primers for amplification of ascomycetous euknr also show a certain degree of unspecificity.
Figure 3.
Phylogenetic placement of soil euknr clones. The phylogenetic tree is based on the partial Molybdenum cofactor (MoCo)-binding domain of EukNR (region A in Figure 1). Phytophthora spp. were used as outgroup, branches lacking environmental sequences were collapsed at common phylogenetic levels. The upper part of the tree is collapsed in (a) and shown fully expanded in (b). Environmental sequences are labelled as follows: genomic clone soil M, genomic clone soil P, genomic clone soil N, cDNA clone soil N. Bootstrap support for branches above 50% is shown. Filled green circle, genomic clone soil M; filled yellow square, genomic clone soil P; filled blue triangle, genomic clone soil N; open blue triangle, cDNA clone soil N.
Table 2. Tentative phylogenetic affiliation and relative abundance (%) of euknr clones amplified from agricultural soil DNA.
| Ordera | Speciesb |
Agricultural soilc |
|||
|---|---|---|---|---|---|
| M | P | Ntot | Nfun | ||
| Pezizomycotina i.s. | Pezizomycotina sp. 1a | 2.1 | |||
| Pezizomycotina sp. 1b | 3.0 | ||||
| Pezizomycotina sp. 2 | 20.1 | ||||
| Pezizomycotina sp. 3 | 1.4 | ||||
| Pezizomycotina sp. 4 | 0.6 | ||||
| Pezizomycotina sp. 5 | 5.3 | ||||
| Pezizomycotina sp. 6 | 3.6 | ||||
| Pezizomycotina sp. 7 | 0.6 | ||||
| Pezizomycotina sp. 8 | 5.3 | ||||
| Capnodiales | Cladosporium herbarum complex | 0.7 | |||
| Devriesia sp. NG_p52 | 2.1 | ||||
| Pleosporales | Pleosporales NG_p49 | 1.7 | 2.9 | ||
| Pleosporales sp. 1 | 0.7 | ||||
| Chaetothyriales | Exophiala salmonis | 1.2 | |||
| Leotiomycetes i.s. | Geomyces sp. | 0.6 | 1.0 | ||
| Helotiales | Tetracladium marchalianum complex | 1.2 | 1.9 | ||
| Tetracladium sp. 1 | 13.0 | ||||
| Tetracladium sp. 2 | 3.5 | ||||
| Sordariomycetes i.s. | Sordariomycetes sp. 1 | 1.7 | 2.9 | ||
| Sordariomycetes sp. 2 | 19.7 | ||||
| Sordariomycetes sp. 3 | 5.6 | ||||
| Sordariomycetes sp. 4 | 29.6 | ||||
| Hypocreales | Fusarium sp. 1 | 2.4 | |||
| Fusarium sp. 2 | 1.2 | 1.9 | |||
| Fusarium sp. 3 | 0.6 | 1.0 | |||
| Hypocreales NG_p26 | 1.2 | 1.9 | |||
| Hypocreales sp. 1a | 0.6 | 1.0 | |||
| Hypocreales sp. 1b | 5.2 | 8.6 | |||
| Microascales | Doratomyces sp. | 21.3 | |||
| Phyllachorales | Gibellulopsis nigrescens | 7.0 | 5.8 | 9.5 | |
| Sordariales | Lasiosphaeriaceae sp. 1 | 7.7 | 4.1 | ||
| Lasiosphaeriaceae sp. 2 | 1.4 | 4.7 | |||
| Lasiosphaeriaceae sp. 3 | 3.0 | ||||
| Lasiosphaeriaceae sp. 4 | 1.7 | 2.9 | |||
| Lasiosphaeriaceae sp. 5 | 16.9 | 17.3 | 28.6 | ||
| Lasiosphaeriaceae sp. 6 | 2.4 | ||||
| Lasiosphaeriaceae WF151 | 4.7 | ||||
| Sordariales sp. 1 | 4.6 | 7.6 | |||
| Trichocladium asperum related 1 | 5.2 | 8.6 | |||
| Trichocladium asperum related 2 | 11.6 | 19.0 | |||
| Trichocladium sp. 1 | 0.7 | ||||
| Xylariales | Arthrinium sp. 1a | 0.6 | |||
| Arthrinium sp. 1b | 4.1 | ||||
| Agaricales | Coprinellus sp. | 0.7 | |||
| Mucoromycotina i.s. | Mucoromycotina sp. 1 | 0.6 | 1.0 | ||
| Peronosporales | Peronosporales sp. 1 | 38.7 | |||
| Stramenopiles i.s. | Stramenopiles sp. 1 | 0.6 | |||
| Number (n)d | 142 | 169 | 173 | 105 | |
| 100.0 | 100.0 | 100.0 | 100.0 | ||
Species are sorted according to class and order, with Ascomycota on the top followed by Basidiomycota (Coprinellus sp. 1), Mucoromycotina and the non-fungal phylum Stramenopiles (Peronosporales sp. 1 and Stramenopiles sp.).
Species epithets like sp. 1, 2 and so on indicate different species of unknown affiliation, where 1a and 1b denote closely related species, whereas epithets like NG_p52 indicates identity at the 96 % threshold level (protein sequences) to insufficiently identified species from the reference database (see Supplementary Table 1).
Agricultural soils from Maissau (M), Purkersdorf (P) and Niederschleinz (N). Data for soil N treatments A, B and C (see Materials and methods) were merged and once calculated for all clones including non-fungal sequences (Ntot) and once for fungal sequences only (Nfung).
Absolute number of clones analysed per library.
As not unusual for fungal communities, the euknr gene pool is very uneven, with the three most dominant species accounting in all three agricultural soils for >50% of the clones.
Fungal euknr gene expression in agricultural soil
Expression of ascomycetous euknr genes was examined in agricultural soil N. As no fungal euknr gene expression could be detected in soil N even after fertilization with KNO3 (from experiments described in Inselsbacher et al., 2010), a second set of experiments was conducted where additional carbohydrates were supplied to the soil microbial community. Soil N was amended with a mixture of cellulose, starch and xylan and fertilized with KNO3. Treatments A, B and C differed by addition of organic fertilizer. Organic fertilizer at the applied dose had no effects on soil pools of NO3–, NH4+ and microbial nitrogen at both sampling time points (data not shown). There was, however, a major effect of carbohydrate addition on soil nitrogen pools: NH4+ content and microbial nitrogen increased slightly, whereas NO3– levels dropped tremendously (Table 3). Accordingly, no fungal euknr gene expression could be detected in soil N without carbohydrate addition, whereas gene expression could consistently be detected in carbohydrate-amended soil N at sampling day 1. At sampling day 6, fungal euknr gene expression could be detected in only 1 out of 13 replicates. Analysis of the fungal community actively expressing euknr at day 1 revealed five species from the ascomycetes and one from the Stramenopiles. The latter was only found in treatment A, but at a very high number. Although four out of the five fungal euknr cDNAs were only detected in one of the three treatments, cDNAs affiliated to Trichocladium asperum related 2 were found in all three treatments. Clone abundances in cDNA libraries were always higher than in genomic libraries. A comparative overview of the results is shown in Table 4 and Figure 3.
Table 3. Nitrogen pools and fungal euknr gene expression in microcosm soils.
| Soil Na | Soil N + CHb | Factorc | P-valued | |
|---|---|---|---|---|
| Day 1e | ||||
| NH4+ nitrogenf | 1.08±0.31 | 2.22±0.55 | × 2 | <0.0001 |
| NO3− nitrogeng | 67.7±12.5 | 0.35±0.062 | ÷200 | <0.0001 |
| Microbial nitrogenh | 45.7±20.5 | 198±28.7 | × 4 | <0.0001 |
| ni | 15 | 13 | ||
| tefj | 15 | 12 | ||
| euknrk | 0 | 9 | ||
| Day 6 | ||||
| NH4+ nitrogen | 0.93±0.31 | 1.85±0.40 | × 2 | <0.0001 |
| NO3− nitrogen | 5.58±3.83 | 0.19±0.063 | ÷30 | <0.0001 |
| Microbial nitrogen | 45.6±17.4 | 172±19.5 | × 4 | <0.0001 |
| n | 10 | 14 | ||
| tef | 10 | 14 | ||
| euknr | 0 | 1 | ||
Agricultural soil from Niederschleinz (N) fertilized with KNO3 equivalent to 36 μg nitrogen per gram dry weight; for experimental details, see Materials and methods. Data for nitrogen pools in soil N without carbohydrate addition were taken from Inselsbacher et al. (2010).
As above, but with addition of carbohydrates (mixture of cellulose, starch and xylan).
Approximate x-fold change in nitrogen pools between experiments without and with carbohydrate addition.
Statistical significance of the differences between treatments (one-way analysis of variance (ANOVA)).
Sampling day.
Plant-available ammonium nitrogen in the soil in μg g−1 dry weight (means±s.d.).
Plant-available nitrate nitrogen in the soil in μg g−1 dry weight (means±s.d.).
Microbial nitrogen in the soil in μg g−1 dry weight (means±s.d.).
Number of microcosms analysed.
Number of microcosms where fungal tef (translation elongation factor 1α) gene expression could be detected.
Number of microcosms where fungal euknr gene expression could be detected.
Table 4. Tentative phylogenetic affiliation of fungal euknr complementary (c)DNA and relative expression levels in microcosms with agricultural soil N (soil from Niederschleinz) after addition of potassium nitrate (KNO3) and carbohydrates.
| |
Aa |
|
|
B |
|
|
C |
|
|
|---|---|---|---|---|---|---|---|---|---|
| gb | cc | c/gd | g | c | c/g | g | c | c/g | |
| Fusarium sp. 2 | 6 (2) | 69 (25) | |||||||
| Lasiosphaeriaceae sp. 4 | 3 (1) | 3 (1) | 3 (1) | 3 (1) | 1.0 | ||||
| Lasiosphaeriaceae sp. 5 | 21 (7) | 51 (24) | 2.4 | 40 (14) | 25 (9) | ||||
| Trichocladium asperum rel. 1 | 14 (5) | 13 (6) | 0.9 | 11 (4) | |||||
| Trichocladium asperum rel. 2 | 21 (7) | 49 (23) | 2.2 | 14 (5) | 87 (39) | 6.1 | 22 (8) | 28 (10) | 1.3 |
| Stramenopiles sp.2 | - (30) | ||||||||
| Ascomycotae | 100 (33) | 100 (47) | 100 (35) | 100 (45) | 100 (36) | 100 (36) |
Treatments differed in application of organic fertilizer (A: no organic fertilizer; B: 0.05% Biovin; C: 0.05% autoclaved Biovin). For further details on the experimental setup, see Materials and methods.
Relative clone abundances (rounded to integer) and absolute clone numbers (in parentheses) in the genomic libraries. Only ascomycetous euknr sequences were considered for calculation of the relative abundances and only species for which euknr cDNA could be found at least in one of the three treatments are shown.
Relative clone abundances (rounded to integer) and absolute clone numbers (in parentheses) in the cDNA libraries. Only ascomycetous euknr sequences were considered for calculation of the relative abundances.
Ratios of relative clone abundances in genomic and cDNA libraries (c/g) were calculated as approximations for expression levels. No relative expression levels were calculated for euknr from Fusarium sp. 2, which was found at the genomic level in treatment B and at the cDNA level in treatment C. Stramenopiles sp. 2 was only found in the cDNA library from treatment A, but in no genomic library.
Number of ascomycetous euknr sequences in the respective libraries, on which the calculation of the relative abundances was based.
Discussion
The current data set for the fungal EukNR phylogenetic tree contains 191 sequences, 82 previously published and 109 newly obtained, and covers all major ascomycete orders found in arable soils (Klaubauf et al., 2010). There are, however, still substantial gaps. No sequences can currently be assigned to soil clone group I, a group of uncultured fungi at the base of the Ascomycota (Porter et al., 2008) that can occur at high frequency, for example, in grassland soil (Klaubauf et al., 2010). As these fungi have not been yet cultivated, it is currently unknown whether or not they can utilize NO3−. From the Taphrinomycotina there are a few completely sequenced genomes, but none of them contains a putative euknr. Within the subphylum Pezizomycotina the class Lecanoromycetes of lichen-forming fungi is not yet represented in the EukNR tree. Based on the available data it seems likely that several events of gains and losses led to today's distribution of euknr in the fungal kingdom: primarily obtained by HGT from an oomycetous donor to an ancestor of the Dikarya (Slot and Hibbett, 2007) or even earlier to a common ancestor of Mucoromycotina and Dikarya (this study, see Supplementary Figure 1), euknr was subsequently frequently lost, mainly in fungi with a yeast-like form in their life cycle. Many species from not only the Saccharomycotina and the Taphrinomycotina, but also basidiomycetous yeasts like Cryptococcus or the dimorphic ascomycete Lecythophora sp. NG_p46 (see Supplementary Table 1) are unable to assimilate nitrate. Good evidence for secondary HGT events is available in two cases: (1) the genera Trichoderma/Hypocrea- and Bionectria-obtained euknr most likely from an ancestor closely related to the Ustilaginaceae (Slot and Hibbett, 2007this study), presumably before they lost their corresponding autochthonous gene, as evidenced by the presence of two copies of euknr in one strain from B. ochroleuca; and (2) the Myxotrichaceae obtained euknr from an ancestor within the Eurotiomycetidae. For other genera with unusual placement of EukNR in the phylogenetic tree (like for example, Doratomyces sp.), bootstrap support is currently lacking to make statements about HGT events and more data are needed. With these exceptions, clustering of most taxa according to established phylogenies (Hibbett et al., 2007) is found in the available EukNR data set. Good correspondence was found between number and position of introns and phylogenetic affiliation, with six introns at conserved positions for all thus far analysed members in the Eurotiales and Onygenales (class Eurotiomycetes) and one intron for all analysed members in the Hypocreales and the Sordariales (class Sordariomycetes). The newly detected HGT event from a Eurotiomycete to an ancestor of the Myxotrichaceae is also supported by conserved intron positions (see Supplementary Figure 3).
As evidenced by the high number of sequences from soil clone libraries that could only be assigned to higher taxonomic levels, expansion of the database of fungal EukNR sequences is fundamental for an accurate affiliation of sequences from environmental samples. The need for a database with good coverage of EukNR from all major fungal lineages is further documented by the common occurrence of HGT events in the evolution of fungal nitrate assimilation. The availability of EukNR sequences beyond the borders of the Molybdenum cofactor-binding domain would moreover be important in data analysis of metagenomic and metatranscriptomic studies.
The relatively high number of agricultural EukNR sequences belonging to the orders Sordariales, Hypocreales and Helotiales is in good agreement with the structure of the fungal communities in these soils as analysed by ITS/LSU (internal transcribed spacer and partial large subunit rRNA gene) libraries (Klaubauf et al., 2010). The dominance of these orders was even more pronounced at the euknr gene expression level in soil N, where all ascomycetous euknr sequences belonged to the Sordariales or Hypocreales. Although many sequences at both the community (=genomic DNA) and gene expression (=cDNA) level could only be loosely affiliated to known families, orders and so on, some sequences could clearly be affiliated to genera or even species. Noteworthy, a set of euknr sequences showing consistently high expression levels in soil N amended with NO3− and carbohydrates was closely related to T. asperum. Fungi related to T. asperum were detected in two out of five investigated agricultural soils, in one of which it was the most abundant species (Klaubauf et al., 2010). although fungi related to T. asperum were not detected in native soil N, they were probably present below detection limit and thrived under the applied conditions, that is, carbohydrate addition and nitrate fertilization. The availability of cultured isolates opens up the possibility to study in more detail the regulation of euknr gene expression of autochthonous fungi in agroecosystems. Regulation of euknr gene expression was thus far mainly studied in response to nitrogen sources (Cove, 1979; Marzluf, 1997; Berger et al., 2006, 2008; Kemppainen et al., 2010; Schinko et al., 2010) and rarely in response to carbon sources (Hynes, 1973; Choudary and Ramananda Rao, 1982). The current data suggest that probably carbon source availability acts as the primary signal. In soil N without addition of carbohydrates, NO3− levels remain high, and expression levels of fungal euknr are undetectable. Fungi are, however, not metabolically inactive under these conditions as evidenced by expression of tef (Table 3). After addition of carbohydrates, fungal euknr gene expression is turned on and NO3− levels drop well below levels of available NH4+. Concomitantly, microbial nitrogen increases, suggesting that at least part of the NO3− is assimilated by fungi for incorporation into biomass. After prolonged incubation (day 6), euknr gene expression levels drop, probably because of the absence of induction by NO3− and increased competition for NO3− by the developing plants (Inselsbacher et al., 2010). In a comparative study, organic farming employing high organic matter inputs had a significantly positive effect on nitrate assimilation by soil microbes compared with conventional farming (Burger and Jackson, 2003). The dominant role of carbon source on fungal euknr gene expression is in good agreement with previous reports that show carbon source dependence of nitrate reductase activity in Candida utilis (Choudary and Ramananda Rao, 1982) and a recent meta-analysis identifying the ratio of organic carbon to NO3− as the main regulator for the fate of NO3− in terrestrial and aquatic ecosystems (Taylor and Townsend, 2010). NO3− reduction is an energy-demanding process requiring strong supply of reduced nicotinamide adenine dinucleotide phosphate from the pentose phosphate pathway in fungi. Consequently, several enzymes of the pentose phosphate pathway are co-regulated with the nitrate assimilation genes in A. nidulans (Hankinson, 1974; Hankinson and Cove, 1974; Schinko et al., 2010).
The presented data will certainly contribute to our understanding of the role of fungi in biogeochemical nitrogen cycles. The availability of partial euknr sequences from a broad taxonomic range of Ascomycota allows incorporation of euknr into functional analytical tools like the GeoChip (He et al., 2007). Furthermore, it will improve phylogenetic affiliation of euknr sequence tags from metatranscriptomic approaches. It must be highlighted that the current data set contains euknr sequences from several orders without completely sequenced genomes like the Botryosphaeriales, Chaetosphaeriales, Chaetothyriales, Diaporthales, Dothideales, Microascales and Xylariales. Future work will further expand the ascomycetous data set and explore groups herein largely neglected like the Basidiomycota or Mucoromycotina. Moreover, the availability of a comprehensive euknr data set will allow the development of efficient tools for quantification of gene expression by real-time PCR techniques in a broad range of environmental samples.
Acknowledgments
This work was supported by Grants LS-05-36 (Nitrogenom) of the Vienna Science, Research and Technology Fund WWTF, S10003-B17 (MicDiF) of the Austrian Science Fund FWF and FP6-FOOD-CT-2006-036297 (RhiBac) of the European Union. Nicko Pritz is acknowledged for his technical assistance in performing growth tests. Publicly available sequences from completed genomes not yet published or deposited at GenBank were obtained through the web portals of the US Department of Energy's Joint Genome Institute (Community Sequencing Program; www.jgi.doe.gov/csp) and the Broad Institute (Fungal Genome Initiative; www.broadinstitute.org/science/projects/fungal-genome-initiative/fungal-genome-initiative). For details, see Supplementary Table 3.
Footnotes
Supplementary Information accompanies the paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alcántara-Hernández RJ, Valenzuela-Encinas C, Zavala-Díaz de la Serna FJ, Rodriguez-Revilla J, Dendooven L, Marsch R. Haloarchaeal assimilatory nitrate-reducing communities from a saline alkaline soil. FEMS Microbiol Lett. 2009;298:56–66. doi: 10.1111/j.1574-6968.2009.01710.x. [DOI] [PubMed] [Google Scholar]
- Ali AH, Hipkin CR. Nitrate assimilation in the basidiomycete yeast Sporobolomyces roseus. J Gen Microbiol. 1985;131:1867–1874. [Google Scholar]
- Ali AH, Hipkin CR. Nitrate assimilation in Candida nitratophila and other yeasts. Arch Microbiol. 1986;144:263–267. [Google Scholar]
- Allen AE, Booth MG, Frischer ME, Verity PG, Zehr JP, Zani S. Diversity and detection of nitrate assimilation genes in marine bacteria. Appl Environ Microbiol. 2001;67:5343–5348. doi: 10.1128/AEM.67.11.5343-5348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arst HN, Cove DJ. Nitrogen metabolite repression in Aspergillus nidulans. Mol Gen Genet. 1973;126:111–141. doi: 10.1007/BF00330988. [DOI] [PubMed] [Google Scholar]
- Berger H, Basheer A, Böck S, Reyes-Dominguez Y, Dalik T, Altmann F, et al. Dissecting individual steps of nitrogen transcription factor cooperation in the Aspergillus nidulans nitrate cluster. Mol Microbiol. 2008;69:1385–1398. doi: 10.1111/j.1365-2958.2008.06359.x. [DOI] [PubMed] [Google Scholar]
- Berger H, Pachlinger R, Morozov I, Goller S, Narendja F, Caddick M, et al. The GATA factor AreA regulates localization and in vivo binding site occupancy of the nitrate activator NirA. Mol Microbiol. 2006;59:433–446. doi: 10.1111/j.1365-2958.2005.04957.x. [DOI] [PubMed] [Google Scholar]
- Bernreiter A, Ramon A, Fernandez-Martinez J, Berger H, Araujo-Bazan L, Espeso EA, et al. Nuclear export of the transcription factor NirA is a regulatory checkpoint for nitrate induction in Aspergillus nidulans. Mol Cell Biol. 2007;27:791–802. doi: 10.1128/MCB.00761-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger M, Jackson LE. Microbial immobilization of ammonium and nitrate in relation to ammonification and nitrification rates in organic and conventional cropping systems. Soil Biol Biochem. 2003;35:29–36. [Google Scholar]
- Campbell WH. Structure and function of eukaryotic NAD(P)H:nitrate reductase. Cell Mol Life Sci. 2001;58:194–204. doi: 10.1007/PL00000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celar F. Competition for ammonium and nitrate forms of nitrogen between some phytopathogenic and antagonistic soil fungi. Biol Control. 2003;28:19–24. [Google Scholar]
- Choudary PV, Ramananda Rao G. Energy-dependence of nitrate reductase induction in Candida utilis. Biochem Biophys Res Commun. 1982;108:1293–1299. doi: 10.1016/0006-291x(82)92140-4. [DOI] [PubMed] [Google Scholar]
- Cove DJ. Genetic studies of nitrate assimilation in Aspergillus nidulans. Biol Rev Camb Philos Soc. 1979;54:291–327. doi: 10.1111/j.1469-185x.1979.tb01014.x. [DOI] [PubMed] [Google Scholar]
- Currah RS. Taxonomy of the Onygenales: Arthrodermataceae, Gymnoascaceae, Myxotrichaceae and Onygenaceae. Mycotaxon. 1985;24:1–216. [Google Scholar]
- Davidson EA, Stark JM, Firestone MK. Microbial production and consumption of nitrate in an annual grassland. Ecology. 1990;71:1968–1975. [Google Scholar]
- Domsch KH, Gams W, Anderson TH.1993Compendium of Soil Fungivol. 1.IHW: Eching [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway JN, Townsend AR, Erisman JW, Bekunda M, Cai Z, Freney JR, et al. Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science. 2008;320:889–892. doi: 10.1126/science.1136674. [DOI] [PubMed] [Google Scholar]
- Glass AD, Britto DT, Kaiser BN, Kinghorn JR, Kronzucker HJ, Kumar A, et al. The regulation of nitrate and ammonium transport systems in plants. J Exp Bot. 2002;53:855–864. doi: 10.1093/jexbot/53.370.855. [DOI] [PubMed] [Google Scholar]
- Gorfer M, Persak H, Berger H, Brynda S, Bandian D, Strauss J. Identification of heavy metal regulated genes from the root associated ascomycete Cadophora finlandica using a genomic microarray. Mycol Res. 2009;113:1377–1388. doi: 10.1016/j.mycres.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Hankinson O. Mutants of the pentose phosphate pathway in Aspergillus nidulans. J Bacteriol. 1974;117:1121–1130. doi: 10.1128/jb.117.3.1121-1130.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankinson O, Cove DJ. Regulation of the pentose phosphate pathway in the fungus Aspergillus nidulans. The effect of growth with nitrate. J Biol Chem. 1974;249:2344–2353. [PubMed] [Google Scholar]
- Hawker KL, Montague P, Kinghorn JR. Nitrate reductase and nitrite reductase transcript levels in various mutants of Aspergillus nidulans: confirmation of autogenous regulation. Mol Gen Genet. 1992;231:485–488. doi: 10.1007/BF00292720. [DOI] [PubMed] [Google Scholar]
- He Z, Gentry TJ, Schadt CW, Wu L, Liebich J, Chong SC, et al. GeoChip: a comprehensive microarray for investigating biogeochemical, ecological and environmental processes. ISMEJ. 2007;1:67–77. doi: 10.1038/ismej.2007.2. [DOI] [PubMed] [Google Scholar]
- Hibbett DS, Binder M, Bischoff JF, Blackwell M, Cannon PF, Eriksson OE, et al. A higher-level phylogenetic classification of the Fungi. Mycol Res. 2007;111:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Huber T, Faulkner G, Hugenholtz P. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics. 2004;20:2317–2319. doi: 10.1093/bioinformatics/bth226. [DOI] [PubMed] [Google Scholar]
- Hynes MJ. The effect of lack of a carbon source on nitrate-reductase activity in Aspergillus nidulans. J Gen Microbiol. 1973;79:155–157. doi: 10.1099/00221287-79-1-155. [DOI] [PubMed] [Google Scholar]
- Inselsbacher E, Hinko-Najera Umana N, Stange FC, Gorfer M, Schüller E, Ripka K, et al. Short-term competition between crop plants and soil microbes for inorganic N fertilizer. Soil Biol Biochem. 2010;42:360–372. [Google Scholar]
- Inselsbacher E, Ripka K, Klaubauf S, Fedosoyenko D, Hackl E, Gorfer M, et al. A cost-effective high-throughput microcosm system for studying nitrogen dynamics at the plant-microbe-soil interface. Plant Soil. 2009;317:293–307. [Google Scholar]
- Jargeat P, Gay G, Debaud JC, Marmeisse R. Transcription of a nitrate reductase gene isolated from the symbiotic basidiomycete fungus Hebeloma cylindrosporum does not require induction by nitrate. Mol Gen Genet. 2000;263:948–956. doi: 10.1007/pl00008695. [DOI] [PubMed] [Google Scholar]
- Jargeat P, Rekangalt D, Verner MC, Gay G, Debaud JC, Marmeisse R, et al. Characterisation and expression analysis of a nitrate transporter and nitrite reductase genes, two members of a gene cluster for nitrate assimilation from the symbiotic basidiomycete Hebeloma cylindrosporum. Curr Genet. 2003;43:199–205. doi: 10.1007/s00294-003-0387-2. [DOI] [PubMed] [Google Scholar]
- Keane TM, Naughton TJ, McInerney JO. MultiPhyl: a high-throughput phylogenomics webserver using distributed computing. Nucleic Acids Res. 2007;35:W33–W37. doi: 10.1093/nar/gkm359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemppainen MJ, Crespo MCA, Pardo AG. fHANT-AC genes of the ectomycorrhizal fungus Laccaria bicolor are not repressed by L-glutamine allowing simultaneous utilization of nitrate and organic nitrogen sources. Environ Microbiol Rep. 2010;2:541–553. doi: 10.1111/j.1758-2229.2009.00111.x. [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, Kawagashira N, Doi K, Kishimoto N, et al. Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- Klaubauf S, Inselsbacher E, Zechmeister-Boltenstern S, Wanek W, Gottsberger R, Strauss J, et al. Molecular diversity of fungal communities in agricultural soils from Lower Austria. Fungal Divers. 2010;44:65–75. doi: 10.1007/s13225-010-0053-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder J. The Yeasts, A Taxonomic Study. North-Holland Publishing Company: Amsterdam; 1970. [Google Scholar]
- Ma LJ, Ibrahim AS, Skory C, Grabherr MG, Burger G, Butler M, et al. Genomic analysis of the basal lineage fungus Rhizopus oryzae reveals a whole-genome duplication. PLoS Genet. 2009;5:e1000549. doi: 10.1371/journal.pgen.1000549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluf GA. Genetic regulation of nitrogen metabolism in the fungi. Microbiol Mol Biol Rev. 1997;61:17–32. doi: 10.1128/mmbr.61.1.17-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty GW, Bremner JM. Regulation of assimilatory nitrate reductase activity in soil by microbial assimilation of ammonium. Proc Natl Acad Sci USA. 1992a;89:453–456. doi: 10.1073/pnas.89.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty GW, Bremner JM. Inhibition of assimilatory nitrate reductase activity in soil by glutamine and ammonium analogs. Proc Natl Acad Sci USA. 1992b;89:5834–5836. doi: 10.1073/pnas.89.13.5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro-Pastor MI, Gonzalez R, Strauss J, Narendja F, Scazzocchio C. The GATA factor AreA is essential for chromatin remodelling in a eukaryotic bidirectional promoter. EMBO J. 1999;18:1584–1597. doi: 10.1093/emboj/18.6.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro-Pastor MI, Strauss J, Ramon A, Scazzocchio C. A paradoxical mutant GATA factor. Eukaryot Cell. 2004;3:393–405. doi: 10.1128/EC.3.2.393-405.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendja F, Goller SP, Wolschek M, Strauss J. Nitrate and the GATA factor AreA are necessary for in vivo binding of NirA, the pathway-specific transcriptional activator of Aspergillus nidulans. Mol Microbiol. 2002;44:573–583. doi: 10.1046/j.1365-2958.2002.02911.x. [DOI] [PubMed] [Google Scholar]
- Nygren CM, Eberhardt U, Karlsson M, Parrent JL, Lindahl BD, Taylor AF. Growth on nitrate and occurrence of nitrate reductase-encoding genes in a phylogenetically diverse range of ectomycorrhizal fungi. New Phytol. 2008;180:875–889. doi: 10.1111/j.1469-8137.2008.02618.x. [DOI] [PubMed] [Google Scholar]
- Porter TM, Schadt CW, Rizvi L, Martin AP, Schmidt SK, Scott-Denton L, et al. Widespread occurrence and phylogenetic placement of a soil clone group adds a prominent new branch to the fungal tree of life. Mol Phylogenet Evol. 2008;46:635–644. doi: 10.1016/j.ympev.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Premakumar R, Sorger GJ, Gooden D. Nitrogen metabolite repression of nitrate reductase in Neurospora crassa. J Bacteriol. 1979;137:1119–1126. doi: 10.1128/jb.137.3.1119-1126.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punt PJ, Strauss J, Smit R, Kinghorn JR, van den Hondel CA, Scazzocchio C. The intergenic region between the divergently transcribed niiA and niaD genes of Aspergillus nidulans contains multiple NirA binding sites which act bidirectionally. Mol Cell Biol. 1995;15:5688–5699. doi: 10.1128/mcb.15.10.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson DJ, Watmough NJ. Inorganic nitrogen metabolism in bacteria. Curr Opin Chem Biol. 1999;3:207–219. doi: 10.1016/S1367-5931(99)80034-9. [DOI] [PubMed] [Google Scholar]
- Sarbhoy AK. Nutritional studies on some members of the mucorales. V. Utilization of different nitrogen compounds. Pathol Microbiol. 1965;28:816–829. [PubMed] [Google Scholar]
- Schinko T, Berger H, Lee W, Gallmetzer A, Pirker K, Pachlinger R, et al. Transcriptome analysis of nitrate assimilation in Aspergillus nidulans reveals connections to nitric oxide metabolism. Mol Microbiol. 2010;78:720–738. doi: 10.1111/j.1365-2958.2010.07363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Siverio JM. Assimilation of nitrate by yeasts. FEMS Microbiol Rev. 2002;26:277–284. doi: 10.1111/j.1574-6976.2002.tb00615.x. [DOI] [PubMed] [Google Scholar]
- Slot JC, Hallstrom KN, Matheny PB, Hibbett DS. Diversification of NRT2 and the origin of its fungal homolog. Mol Biol Evol. 2007;24:1731–1743. doi: 10.1093/molbev/msm098. [DOI] [PubMed] [Google Scholar]
- Slot JC, Hibbett DS. Horizontal transfer of a nitrate assimilation gene cluster and ecological transitions in fungi: a phylogenetic study. PLoS ONE. 2007;2:e1097. doi: 10.1371/journal.pone.0001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stitt M, Muller C, Matt P, Gibon Y, Carillo P, Morcuende R, et al. Steps towards an integrated view of nitrogen metabolism. J Exp Bot. 2002;53:959–970. doi: 10.1093/jexbot/53.370.959. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S.2011MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods Mol Biol Evol(in press). [DOI] [PMC free article] [PubMed]
- Taylor PG, Townsend AR. Stoichiometric control of organic carbon-nitrate relationships from soils to the sea. Nature. 2010;464:1178–1181. doi: 10.1038/nature08985. [DOI] [PubMed] [Google Scholar]
- Tsuneda A, Currah RS. Ascomatal morphogenesis in Myxotrichum arcticum supports the derivation of the Myxotrichaceae from a discomycetous ancestor. Mycologia. 2004;96:627–635. [PubMed] [Google Scholar]
- Wang Z, Binder M, Schoch CL, Johnston PR, Spatafora JW, Hibbett DS. Evolution of helotialean fungi (Leotiomycetes, Pezizomycotina): a nuclear rDNA phylogeny. Mol Phylogenet Evol. 2006a;41:295–312. doi: 10.1016/j.ympev.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Wang Z, Johnston PR, Takamatsu S, Spatafora JW, Hibbett DS. Toward a phylogenetic classification of the Leotiomycetes based on rDNA data. Mycologia. 2006b;98:1065–1075. doi: 10.3852/mycologia.98.6.1065. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Wong KH, Hynes MJ, Davis MA. Recent advances in nitrogen regulation: a comparison between Saccharomyces cerevisiae and filamentous fungi. Eukaryot Cell. 2008;7:917–925. doi: 10.1128/EC.00076-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.