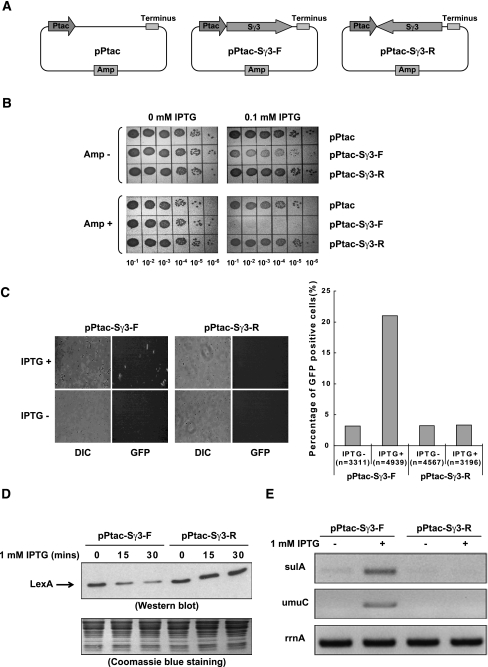
Figure 1.
Transcription through a plasmid-borne Sγ3 region in its physiological orientation induces cell growth defect, plasmid loss, and SOS damage response in E. coli. (A) Schematic diagram of pPtac, pPtac-Sγ3-F, and pPtac-Sγ3-R plasmids. Positions of the Ptac promoter, transcription terminus, Sγ3 region, and amp resistance gene are indicated. (B) Determination of the viability of transformants in the presence or absence of 0.1 mM IPTG. E. coli K-12 BW25113 cells were transformed by pPtac, pPtac-Sγ3-F, or pPtac-Sγ3-R plasmids, respectively. A single colony from each transformant was inoculated into LB with 100 μg/mL Amp. Tenfold dilutions of overnight cultures were grown on LB plates containing 0.1 mM IPTG and/or 100 μg/mL Amp. (C) Sγ3 transcription in its physiological orientation activates the SOS-inducible sulA promoter. pPtac-Sγ3-F or pPtac-Sγ3-R was transformed into an E. coli SMR8379 strain in which a chromosomally located gfp gene is under the control of a SOS-inducible sulA promoter. Transformants were cultured in the presence or absence of 1 mM IPTG for 30 min and allowed to recover for 2 h. Expression of GFP was monitored by fluorescent microscope. Representative images are presented. Quantification is graphically shown in the right panel. The number of cells counted for each sample is indicated. (DIC) Differential interference contrast image; (GFP) GFP fluorescence image. (D,E) E. coli K-12 BW25113 cells were transformed with pPtac-Sγ3-F or pPtac-Sγ3-R plasmids, respectively. Transformants were grown in the presence of 1 mM IPTG for the indicated times and allowed to recover for 30 min. (D, top panel) Cell lysates were assayed for LexA by Western blot. (Bottom panel) The amount of proteins loaded onto each lane of the SDS-PAGE gel was monitored by Coomassie blue staining. (E) The mRNA levels of the sulA (top panel), umuC (middle panel), and rrnA (bottom panel) genes were determined by RT–PCR.