Abstract
Patterning of different cell types in embryos is a key mechanism in metazoan development. Communities of microorganisms, such as colonies and biofilms also display patterns of cell types. For example, in the yeast S. cerevisiae, sporulated cells and pseudohyphal cells are not uniformly distributed in colonies. The functional importance of patterning and the molecular mechanisms that underlie these patterns are still poorly understood.
One challenge with respect to investigating patterns of cell types in fungal colonies is that unlike metazoan tissue, cells in colonies are relatively weakly attached to one another. In particular, fungal colonies do not contain the same extensive level of extracellular matrix found in most tissues . Here we report on a method for embedding and sectioning yeast colonies that reveals the interior patterns of cell types in these colonies. The method can be used to prepare thick sections (0.5 μ) useful for light microscopy and thin sections (0.1 μ) suitable for transmission electron microscopy. Asci and pseudohyphal cells can easily be distinguished from ovoid yeast cells by light microscopy , while the interior structure of these cells can be visualized by EM.
The method is based on surrounding colonies with agar, infiltrating them with Spurr's medium, and then sectioning. Colonies with a diameter in the range of 1-2 mm are suitable for this protocol. In addition to visualizing the interior of colonies, the method allows visualization of the region of the colony that invades the underlying agar.
Protocol
1. Colony Isolation and Fixation
Incubate 300 colonies on agar medium for the indicated time (an isolated colony should be 1-2 mm in diameter).
Remove colony (face up) and underlying medium using a narrow spatula.
Place several drops of 2% agar 42°C on a microscope slide using 1 mL pipetman tip and immediately place colony on agar face up before it solidifies.
Place several drops of 2% agar 42°C on colony and allow to solidify.
Wear gloves for all steps through the remainder of this protocol
Trim block with razor blade and place them in a 3.5 mL borosilicate screw-cap vial containing 2% paraformaldehyde/2% glutaraldehyde fixative for 7 days at 4°C.
2. Washes and Osmium Treatment
After about 1 week, wash agar blocks on ice under chemical fume hood by incubating twice for 15 minutes with enough 0.15M sodium cacodylate (pH 7.2) to completely cover the colony (approximately 1.5 mL) then twice more for 5 minutes with the same volume of 1X OS buffer (100 mM KH2PO4 10 mM MgCl2, pH 6.0).
Add the same volume of 1% OsO4 [2 mL OsO4 (2%) + 2 mL 2x OS Buffer] to vials (on ice) to cover the agar blocks under chemical fume hood for 1 hour then wash twice with the same amount of 1X OS buffer for 10 minutes each.
Leave vials overnight at 4°C in 1X OS buffer.
Fixative and all washes in this step are disposed of as hazardous waste. All pipettes containing OsO4 were rinsed 3X with vegetable oil.
3. Wash and Dehydration
The next morning wash agar blocks twice on ice with the same volume as above of cold water for 10 minutes each using Pasteur pipette.
Wash sequentially with the same volume of 25%,50%,75%,95% ethanol for 10 minutes each followed by 2 washes with 100% ethanol for 10 minutes. Leave overnight at 4°C in 100% ethanol.
All washes in this step are disposed of as hazardous waste.
4. Spurr's Preparation
The following morning (as early as possible) weigh the following solutions (EM Sciences) into a disposable plastic beaker under a fume hood to prepare Spurr's reagent. For all subsequent work using this reagent, continue to use a fume hood. (ERL 4221 solution 5 grams) (DER 736 solution 4 grams) (NSA solution 13 grams)
mix these solutions (slowly) for 20 minutes in hood using a stir bar
Add DMAE solution 0.15 grams and stir 20 minutes as above.
Degas above mixture for 1-2 hrs in hood.
5. Spurr's Infiltration
As soon as Spurr's is made wash agar blocks 5 times with same volume as above of 100% room temp ethanol for 10 minutes each. After the last ethanol wash remove the ethanol using a Pasteur pipette so that the remaining ethanol just covers the agar blocks.
Add approximately an equal amount of Spurr's reagent (approximately 0.5 mL) and rotate vials on wheel for 15 minutes at room temperature and then allow to stand for 30 minutes. After 30 minutes remove the solution completely using a Pasteur pipette, add Spurr's to cover agar block (approximately 1.5 mL), rotate vials on wheel for 15 minutes at room temperature and allow to stand for 30 minutes. Repeat the Spurr's replacement, rotation, and incubation 3 more times.
After the final 30 minutes, remove Spurr's reagent and add fresh Spurr's to cover the blocks. Allow the agar blocks to stand at room temp for 4 hrs.
Replace the Spurr's and rotate overnight.
In the morning replace Spurr's and leave rotating until the following day.
The next morning place Spurr's in numbered silicone molds (Fisher Scientific) just to cover the bottom of molds then add agar blocks to the molds. Incubate at 60°C for 4 hours
After 4 hrs, top off the molds, with more Spurr's and incubate at 60°C overnight.
If after removing the blocks from molds the blocks are still slightly flexible, return to the molds and incubate an additional 2 days
6. Sectioning
A Leica Ultracut S microtome was used to cut 0.5 μ thick sections for light microscopy and 0.1 μ thin sections for transmission EM.
For light microscopy, as sections were cut, they were placed on a drop of dH2O, and dried on a 52°C heat block. Dried sections were then stained with a drop of 1.0 % toluidine blue, 1% Sodium Borate for 5-15 seconds. After staining, the sections were immediately washed under a stream of water, dried, covered in mounting media (KPL), and a cover slip sealed over the sample.
7. Representative Results:
The broad relevance of measuring pattern formation in yeast and other microorganisms have been reviewed previously 1. Of particular note is the increased functionality of organized microbial colonies relative to disorganized communities.
The described method is a modification of a method for embedding larger specialized colonies 2, and a short description of our modifications has been published 3.
An example of a light micrograph of a section through the center of a colony of wild S. cerevisiae yeast colony is shown in Figure 1. Asci, pseudohyphae and ovoid yeast are easily distinguishable, and the region of the colony invading the underlying agar is also apparent.
An example of an electron micrograph of a laboratory strain of yeast (W303 background) is shown in Figure 2. The image is from a region of the colony containing a high frequency of sporulated cells.
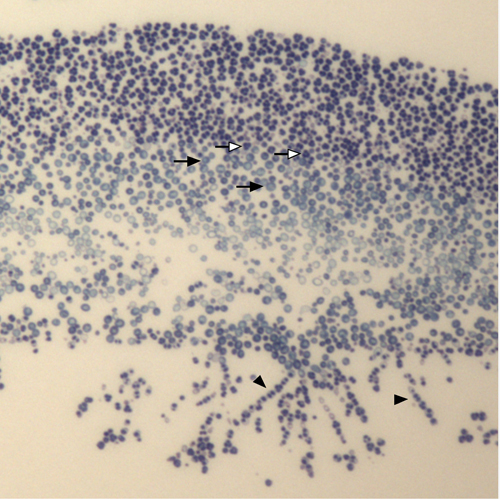 Figure 1. Light microscopy of a wild yeast colony. This yeast (YPS133) was isolated recently from tree exudates 4. The colony was incubated for 6 days on YNA medium 5 prior to sectioning. Open arrows indicate representative asci, filled arrows indicate representative ovoid vegetative cells, filled arrowheads indicate chains of elongated cells (pseudohyphae) and asci.
Figure 1. Light microscopy of a wild yeast colony. This yeast (YPS133) was isolated recently from tree exudates 4. The colony was incubated for 6 days on YNA medium 5 prior to sectioning. Open arrows indicate representative asci, filled arrows indicate representative ovoid vegetative cells, filled arrowheads indicate chains of elongated cells (pseudohyphae) and asci.
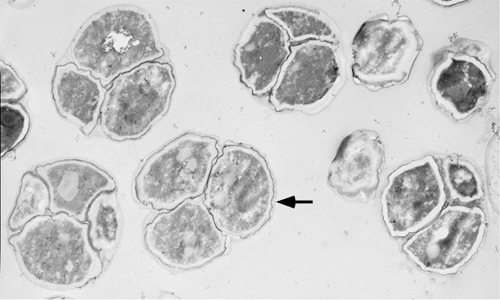 Figure 2. Electron microscopy of a laboratory yeast strain. The colony of SH1020 (W303 background) was incubated for 6 days on YNA medium prior to sectioning 3. Image shows a region of the section with a high frequency of asci. An arrow indicates the bi-layer structure of the spore wall.
Figure 2. Electron microscopy of a laboratory yeast strain. The colony of SH1020 (W303 background) was incubated for 6 days on YNA medium prior to sectioning 3. Image shows a region of the section with a high frequency of asci. An arrow indicates the bi-layer structure of the spore wall.
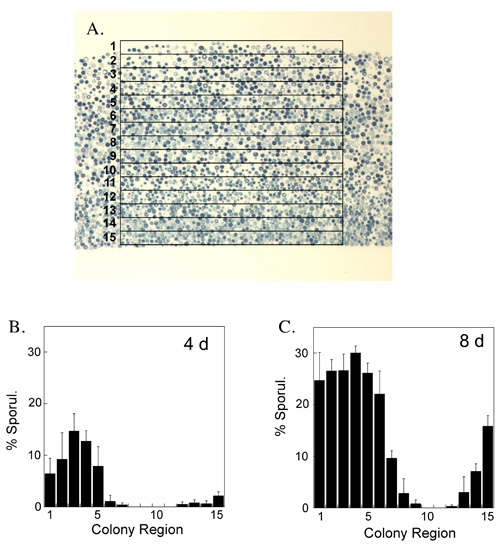 Figure 3. Quantification of the distribution of sporulated cells in colonies: A) a grid containing 15 adjoining rectangles stacked along their long side is superimposed on the central region of an image of a colony section. The grid is scaled, while keeping its proportions constant, to just covers the top and bottom of the colony. (B & C) The fraction of sporulated cells in each rectangle relative to the total cells in that rectangle is determined by visual inspection of images from 4-day old (B) and 8-day old (C) colonies. For example, the rectangle at the top of the colony (labeled "1") corresponds to the left side of the graph. Mean of 4 colonies is shown; the error bars display the std. error.
Figure 3. Quantification of the distribution of sporulated cells in colonies: A) a grid containing 15 adjoining rectangles stacked along their long side is superimposed on the central region of an image of a colony section. The grid is scaled, while keeping its proportions constant, to just covers the top and bottom of the colony. (B & C) The fraction of sporulated cells in each rectangle relative to the total cells in that rectangle is determined by visual inspection of images from 4-day old (B) and 8-day old (C) colonies. For example, the rectangle at the top of the colony (labeled "1") corresponds to the left side of the graph. Mean of 4 colonies is shown; the error bars display the std. error.
Discussion
The method presented reveals the interior structures of colonies. Because the method is effective in determining patterns of cell types in a range of S. cerevisiae strains with different colony morphologies, and also in a related species S. paradoxus 5, the method is also likely to work on a wide range of fungi and other microorganisms.
One critical step for the success of the method is to ensure that the entire colony, including the top of the colony, is encased in agar throughout the protocol. Whether colonies are completely encased in the agar can be determined after sectioning for light microscopy and staining with toluidine blue. When stained sections are viewed by light microscopy, the agar medium is stained slightly darker than the embedding medium. Because agar shrinks during the dehydrations steps, in order to recover the entire colony be sure to: 1) add 4-5 drops of agar to reliably cover the colony in step 1.2, and 2) trim the agar only on the sides, not on the top and bottom, and only trim enough to allow it to fit within molds in step 1.5.
A second stage of the protocol that is critical for optimal sections involves the hardness of the block of embedding medium. Typically blocks are examined after overnight incubation, by removing them from the molds. If the blocks are still flexible when held between both hands and flexed, they are probably not hard enough for sectioning. In this case they are returned to the incubator at the same temperature for an additional 48 hours and retested as above.
The significance of being able to detect patterns of cell types within microbial colonies is that these patterns likely reflect a functional organization of cells into communities. Indeed, microbial patterning may reflect ancient and fundamental mechanisms by which organisms of the same species interact. Together with methods for monitoring patterns of gene expression in colonies 3,6, the ability to detect patterns of cell differentiation in these communities can help to test potential mechanisms of microbial pattern formation.
Disclosures
No conflicts of interest declared.
Acknowledgments
Research was funded by NIH 1R15GM094770.
References
- Kimmel AR, Firtel RA. Breaking symmetries: regulation of Dictyostelium development through chemoattractant and morphogen signal-response. Curr Opin Genet Dev. 2004;14:540–540. doi: 10.1016/j.gde.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Palkova Z, Vachova L. Life within a community: benefit to yeast long-term survival. FEMS Microbiol Rev. 2006;30:806–806. doi: 10.1111/j.1574-6976.2006.00034.x. [DOI] [PubMed] [Google Scholar]
- Shapiro J, Vachova L. The significances of bacterial colony patterns. Bioessays. 1995;17:597–597. doi: 10.1002/bies.950170706. [DOI] [PubMed] [Google Scholar]
- Shapiro J, Vachova L. Thinking about bacterial populations as multicellular organisms. Annu Rev Microbiol. 1998;52:81–81. doi: 10.1146/annurev.micro.52.1.81. [DOI] [PubMed] [Google Scholar]
- Engelberg D. Multicellular stalk-like structures in Saccharomyces cerevisiae. J Bacteriol. 1998;180:3992–3992. doi: 10.1128/jb.180.15.3992-3996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz R, Shinder V, Engelberg D. Anatomical analysis of Saccharomyces cerevisiae stalk-like structures reveals spatial organization and cell specialization. J Bacteriol. 2001;183:5402–5402. doi: 10.1128/JB.183.18.5402-5413.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccirillo S. The Rim101p/PacC pathway and alkaline pH regulate pattern formation in yeast colonies. Genetics. 2010;184:707–707. doi: 10.1534/genetics.109.113480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski PD, Dombrowski PG, Fingerman E. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 2002;1:299–299. doi: 10.1111/j.1567-1364.2002.tb00048.x. [DOI] [PubMed] [Google Scholar]
- Piccirillo S, Honigberg SM. Sporulation patterning and invasive growth in wild and domesticated yeast colonies. Res Microbiol. 2002;161:390–390. doi: 10.1016/j.resmic.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vachova L. Architecture of developing multicellular yeast colony: spatio-temporal expression of Ato1p ammonium exporter. Environ Microbiol. 2009 doi: 10.1111/j.1462-2920.2009.01911.x. [DOI] [PubMed] [Google Scholar]


