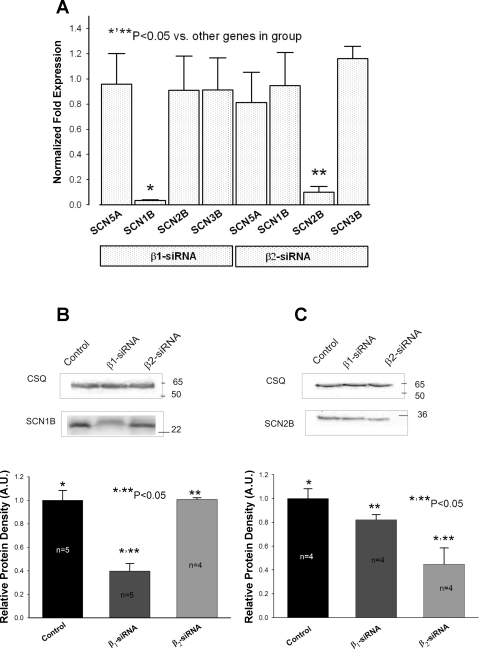
Fig. 1.
Efficacy of virally delivered Navβ1-short interfering RNA (siRNA) and Navβ2-siRNA to post-transcriptionally silence SCN1B and SCN2B genes in ventricular cardiac myocytes from dogs with chronic heart failure (HF). A: SCN5A, SCN1B, SCN2B, and SCN3B mRNA expression levels detected by the real-time PCR and normalized to GAPDH mRNA and to the respective mRNA levels detected in cells transfected with nonsilencing siRNA (2−ΔΔCt method) for each gene. *,**Significantly reduced mRNA level compared with other genes in the group (P < 0.05, ANOVA followed by Bonferroni's post hoc test). Data gathered from 4 to 7 transfections. B and C, top: representative Western blot of Navβ1 (SCN1B) and Navβ2 (SCN2B) subunits (bottom bands) and housekeeping protein calsequestrin (CSQ; top bands) for normalization in cells treated with virally delivered siRNA. Control stands for nonsilencing siRNA. B and C, bottom: bar graphs, in arbitrary units (AU), of the relative protein density, which demonstrates dramatic reduction of Navβ1 or Navβ2 in response to the transfection with concomitant siRNA when normalized to the CSQ, and then normalized to the quantity of the immunoreactive protein in cells transfected with nonsilencing siRNA. Membrane proteins were isolated from at least 3 transfections for each condition (n is shown at the bars). Control in B and C stands for the control, nonsilencing siRNA. *,**Protein levels that were significantly reduced compared with the control (P < 0.05, P < 0.05, ANOVA followed by Bonferroni's post hoc test). In this and in the rest of the figures Navβ1 and Navβ2 were abbreviated to β1 and β2, respectively.