Abstract
Background and Aims
The cool temperate rainforests of Australia were much reduced in range during the cold and dry glacial periods, although genetic evidence indicates that two key rainforest species, Nothofagus cunninghamii and Tasmannia lanceolata, survived within multiple locations and underwent only local range expansions at the end of the Last Glacial. To better understand the glacial response of a co-occurring but wind-dispersed and less cold-tolerant rainforest tree species, Atherosperma moschatum, a chloroplast phylogeographic study was undertaken.
Methods
A total of 3294 bp of chloroplast DNA sequence was obtained for 155 samples collected from across the species' range.
Key Results
The distribution of six haplotypes observed in A. moschatum was geographically structured with an inferred ancestral haplotype restricted to Tasmania, while three non-overlapping and endemic haplotypes were found on the mainland of south-eastern Australia. Last glacial refugia for A. moschatum are likely to have occurred in at least one location in western Tasmania and in Victoria and within at least two locations in the Great Dividing Range of New South Wales. Nucleotide diversity of A. moschatum was lower (π = 0·00021) than either N. cunninghamii (0·00101) or T. lanceolata (0·00073), and was amongst the lowest recorded for any tree species.
Conclusions
This study provides evidence for past bottlenecks having impacted the chloroplast diversity of A. moschatum as a result of the species narrower climatic niche during glacials. This hypothesis is supported by the star-like haplotype network and similar estimated rates of chloroplast DNA substitution for A. moschatum and the two more cold tolerant and co-occurring species that have higher chloroplast diversity, N. cunninghamii and T. lanceolata.
Keywords: Atherosperma moschatum, A. moschatum subsp. integrifolium, Atherospermataceae, bottleneck, comparative phylogeography, glacial refugia, Nothofagus cunninghamii, Pleistocene glacials, south-eastern Australia, Tasmannia lanceolata, cool temperate rainforest
INTRODUCTION
Understanding the ability of species to occupy newly available habitat is fundamental to predicting how species respond to environmental change. The dramatic environmental changes that have occurred since the Last Glacial Maximum (LGM) approx. 18 000 years ago provide a model for developing such an understanding. Worldwide, most areas of temperate forest vegetation are believed to have been established since the onset of favourable climates approx. 13 000 years ago through expansion from refugia in which they survived the cold and, in many regions, dry LGM conditions. The rate and the degree of colonization of the newly available habitat is largely considered to have been determined by individual responses of species (Davis, 1976; Huntley and Birks, 1983; Huntley and Webb, 1989) related to the number and location of glacial refugia, the individual fitness of species, and to life-history traits, including the mechanism of seed dispersal.
Australia's cool temperate rainforests provide an interesting system in which to investigate these processes. These evergreen, closed-canopy forests are widespread but discontinuously distributed within the continent's mountainous south-east corner and the island of Tasmania (Hill et al., 1988). These forests are species poor, especially in trees, but contain many lineages with fossil histories extending back to the mid-Tertiary (30 mya) or earlier (Carpenter and Jordan, 1997; Hill, 2004; Sniderman and Jordan, 2011). Furthermore, fossil evidence indicates that almost all contemporary species have been present at least since the Early Pleistocene (2·5 million years to 788 000 years ago), and therefore have remained relatively unchanged through many, or perhaps all, of the glacial-interglacial cycles of the last few million years (Jordan, 1997). However, only recently has clear evidence for the location of glacial refugia and the geographic extent of post-glacial recovery for individual species of the cool temperate rainforest biome become available. Chloroplast phylogeographic evidence has shown that at least some of the species were highly resilient in response to the major climate changes of the Pleistocene. Chloroplast phylogeographies provide clear evidence for geographic stasis and glacial survival within multiple refugia for three species in Australian cool temperate rainforests: the gravity-dispersed dominant tree species, Nothofagus cunninghamii (Worth et al., 2009) and Eucalyptus regnans (Nevill et al., 2010); and the fleshy-fruited shrub, Tasmannia lanceolata (Worth et al., 2010). Evidence from a fourth species, the gravity- or water-dispersed conifer Lagarostrobos franklinii (Clark and Carbone, 2008), is sparser but is also consistent with stasis.
This study investigates the chloroplast phylogeography of Atherosperma moschatum (Atherospermataceae), a tree species for which there are several reasons to suspect a divergent response to the Pleistocene climatic perturbations compared with other Australian cool temperate rainforest species. Atherosperma moschatum has the most extensive latitudinal range of any Australian cool temperate rainforest woody species and has plumose, wind-dispersed seeds, that have been shown to have efficient dispersal over 150 m distance (Hickey et al., 1982). The extensive geographical distribution of A. moschatum, particularly in contrast to the gravity-dispersed N. cunninghamii (a species that co-occurs extensively with A. moschatum in Tasmania and the highlands of Victoria), has been considered to have been facilitated by the greater ability of the species to disperse during the Holocene over areas of unsuitable habitat (Hickey et al., 1982; Read and Busby, 1990; Neyland and Brown, 1993). For example, the occurrence of A. moschatum in eastern Gippsland of Victoria and in some small, topographically protected sites in the dry eastern half of Tasmania, where N. cunninghamii is absent, has been considered to be a consequence of the ability of A. moschatum to reach suitably wet isolated habitat via long-distance dispersal (Gilbert, 1959; Howard and Ashton, 1973; Neyland and Brown, 1993). Furthermore, physiological evidence suggests that A. moschatum is the least cold tolerant of all Australian cool temperate rainforest trees (Read and Hill, 1989; Read and Busby, 1990; Feild and Brodribb, 2001) and, as such, is a less likely candidate to have withstood glacial conditions in multiple regions. In addition, although drought tolerance has been investigated in only a small number of Australian cool temperate rainforest trees, A. moschatum was the most vulnerable to leaf hydraulic conductance failure during drought compared with three other widespread trees, N. cunninghamii, Pittosporum bicolor and T. lanceolata (Blackman et al., 2010). Currently, there is little evidence available to discern the roles of either multiple glacial refugia or dispersal in explaining the widespread distribution of A. moschatum, largely because the fossil record is uninformative on this matter. Fossil pollen of A. moschatum is rare, with the only LGM records being from some parts of western Tasmania (Macphail and Colhoun, 1985; Colhoun et al., 1999; Hopf et al., 2000), although trace quantities have been found in the Australian Alps at approx. 32 000 years before present (Kershaw et al., 2007; see Supplementary Data Fig. S1, available online). A previous genetic study of 22 populations based on isozymes suggested that populations in south-eastern Tasmania were closely related, while the three Victorian populations had different affinities, either nested within Tasmanian variation or closest to the most northern populations in New South Wales (NSW) (Shapcott, 1994). These most northern populations of A. moschatum in the Blue Mountains and Barrington Tops regions (Figs 1 and 2) were the most distinctive in the species. This finding suggests long term isolation of these northernmost populations (Floyd, 1990; Shapcott, 1994). Populations of the species in these two areas have been classified as a separate subspecies, subsp. integrifolium, mainly based on leaf morphology (Schodde, 1969; Foreman and Whiffen, 2007). A later study of leaf oil chemistry in the genus broadened the range of A. moschatum subsp. integrifolium to include a previously unstudied population at Monga approx. 200 km to the south of the Blue Mountains, NSW (Brophy et al., 2009).
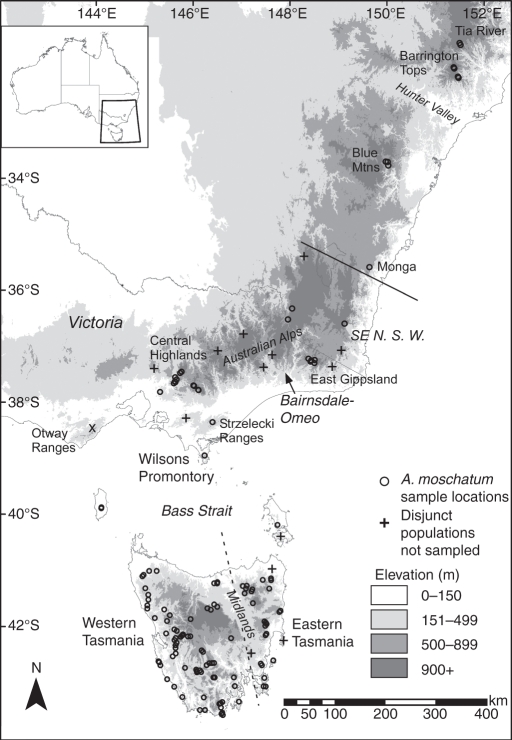
Fig. 1.
Sample locations of Atherosperma moschatum (black circles, in some cases representing two or three plants; Fig. 2 shows the species' distribution). Some major geographic features, including areas of dry unsuitable habitat, are labelled in italics. The continuous line indicates the southernmost known extent of the subspecies, Atherosperma moschatum subsp. integrifolium. The broken line indicates the demarcation of eastern and western Tasmania through the dry Midlands of the island. The × indicates where the species occurred in the Otway Ranges until at least the 1860s and where it is now extinct (Lunt, 1992).
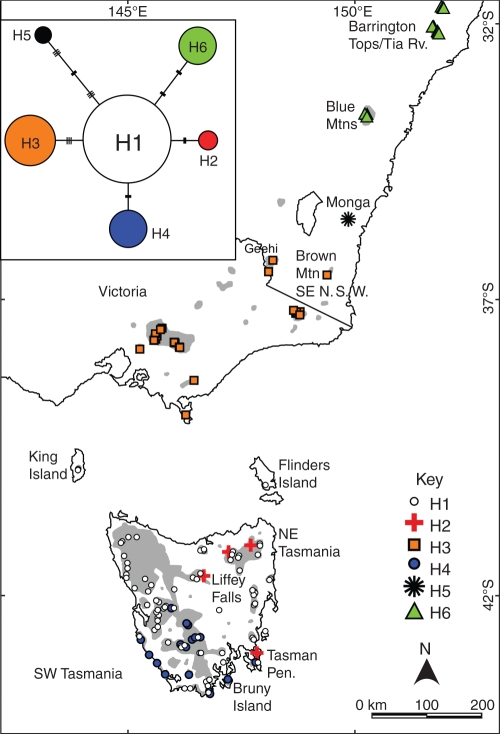
Fig. 2.
The inset shows the median-joining network of the six haplotypes observed across the range of Atherosperma moschatum. The area of the circles in the haplotype network is proportional to the frequency of each haplotype. Lengths of lines connecting each haplotype are proportional to the number of character differences between them. Single nucleotide polymorphisms (SNPs) are indicated by solid bars and simple sequence repeats (SSRs) are indicated by three stacked lines. The main figure shows the distribution of the six haplotypes observed in A. moschatum (H1–H6). Grey areas indicate the natural distribution of the species [derived from the Natural Values Atlas (Department of Primary Industries and Water, Tasmanian Government), Australia's Virtual Herbarium (www.ersa.edu.au/avh/), Floyd (1989, 1990), Coyne (2001) and Ashton (2000)].
This study aimed to identify the responses of A. moschatum to past climatic perturbations by investigating the species' range-wide chloroplast DNA (cpDNA) phylogeography. Specifically, this study assessed whether the species responded to the Pleistocene glacials via local expansion from multiple glacial refugia or, alternatively, with widespread colonization during the Holocene via wind dispersal.
MATERIALS AND METHODS
The study species
Atherosperma moschatum Labill. (southern or black sassafras) is endemic to mainland south-eastern Australia and the island of Tasmania, and is the only species of its genus. This dioecious evergreen tree species can reach 45 m in height (Curtis, 1993). In Tasmania, A. moschatum is common from sea-level to approx. 900 m a.s.l., but does occasionally occur as a shrub at higher altitudes (Read and Hill, 1988a). On mainland Australia, the species is confined to mountainous areas, above 340 m a.s.l. in Victoria, 700–1000 m a.s.l. in south-eastern NSW, and generally above 1200 m a.s.l. in the northern part of its range at Barrington Tops and Tia River (Floyd, 1990), where the species has a very localized occurrence (Read and Hill, 1989). Across its 12° range in latitude (Read and Hill, 1988a), A. moschatum occurs within cool temperate rainforest, often as a subdominant to Nothofagus species. However, it is the sole cool temperate rainforest tree in parts of eastern Tasmania (Kirkpatrick, 1981; Neyland and Brown, 1993), in the Dandenong Range of Victoria (Howard and Ashton, 1973) and in localized parts of the Australian Alps (Gellie, 2005). It can also extend into the sub-canopy of wet eucalyptus forest (Gilbert, 1959; Ashton, 2000). In one location, the Otway Ranges of western Victoria (Fig. 1) where N. cunninghamii is the sole dominant rainforest tree, A. moschatum was present in the region during the Holocene until it went extinct after European settlement, with the last record of the species in the early 1860s (Lunt, 1992; McKenzie and Kershaw, 1997).
Sampling
Leaves were sampled from 155 A. moschatum individuals representative of virtually the whole species' geographic range (Fig. 1). In regions where the distribution of A. moschatum was more or less continuous (e.g. western Tasmania and the central highlands of Victoria) individuals were collected a minimum of approx. 1 km apart to maximize geographic coverage. Within some small and disjunct populations two or three samples were taken where ever possible. In total, 20 individuals of the northern subspecies, A. moschatum subsp. integrifolium (A.Cunn. ex Tul.) Schodde (Schodde, 1969; Foreman and Whiffen, 2007; Brophy et al., 2009), and 135 individuals of the southern subspecies, A. moschatum subsp. moschatum, were sampled (see Supplementary Data Table S1).
For use as outgroups, one species each of three other genera of Atherospermataceae were sampled. These were the Chilean endemic Laurelia sempervirens (Ruiz & Pav.) Tul. (grown in a private collection by Ken Gillanders, southern Tasmania), Nemuaron vieillardii (Baill.) Baill. (from Mont Dzumac, New Caledonia) and Daphnandra tenuipes Perkins (from the Border Ranges National Park, north-eastern NSW).
Molecular methods
Total genomic DNA was extracted from 0·25 g of adult leaves using the Qiagen DNeasy Plant Mini Kit (Qiagen Pty Ltd, Vic, Australia). DNA quantity and quality were assessed by agarose gel electrophoresis with ethidium bromide staining and comparison with a standard molecular weight marker (lambda HindIII).
Eight primer pairs were trialled in a preliminary search for cpDNA variation via direct sequencing within 24 samples representative of most of the species range (for fragment names, primer pairs and references see Supplementary Data Table S2). PCR conditions were as follows: for atpB-1-rbcL-1, an initial 4 min at 94 °C, followed by 35 cycles of 45 s at 94 °C, 1 min and 15 s at 55 °C and an extension for 1 min and 15 s at 72 °C, and a final extension step for 10 min at 72 °C; for K1-matK1 and matK6-K2, an initial 4 min at 94 °C, followed by 35 cycles of 45 s at 94 °C, 1 min at 51 °C and extension for 1·5 min at 72 °C, and a final extension step for 10 min at 72 °C; for psbM-trnD, petN-psbM and a-b reactions, an initial 4 min at 94 °C, followed by 35 cycles of 1 min at 94 °C, 2 min at 51 °C and extension for 2 min at 72 °C, and a final extension step for 10 min at 72 °C; for e-f and c-d, an initial 1 min at 95 °C, followed by 35 cycles of 1 min at 95 °C, 1 min at 50 °C and extension for 45 s at 72 °C, and a final extension step for 7 min at 72 °C. Quality of PCR products was assessed by gel electrophoresis on 1·2 % agarose gels and staining with ethidium bromide. In preparation for DNA sequencing, PCR products were purified using the Qia-Quick PCR purification kit (Qiagen Pty Ltd). DNA sequencing was performed in one direction using the forward primers K1, matK6, psbM2, a and c except for the atpB-rbcL and petN-psbM fragments which were sequenced in both directions.
The trnL-trnF fragment did not amplify, a finding also observed by Renner et al. (2000) for A. moschatum. Since no variation was observed within the psbM-trnD and atpB-rbcL fragments, only the K1-matK1, matK6-K2, petN-psbM, trnT-trnL and the trnL intron fragments were sequenced for all 155 samples.
Sequencing reactions were performed in an MJ Research PTC-225 thermal cycler using ABI prism Bigdye terminator v 3·0 cycle sequencing kits (Applied Biosystems, CA, USA) with AmpliTaq DNA polymerase (Applied Biosystems) and fragment separation was undertaken on a 3730xl DNA analyser (Applied Biosystems). Sequences were aligned using Sequencher 4·5 (Gene Codes Corporation, MI, USA), and checked by eye for incorrect base calls, single nucleotide polymorphisms (SNPs), DNA insertions and deletions (indels) and simple sequence repeat (SSR) variations.
Haplotype network and phylogenetic analyses
For all A. moschatum haplotypes, a median-joining network, with equal weighting of all characters, was constructed using Network 4·5·0·2 (Bandelt et al., 1999). To polarize the haplotype relationships a phylogenetic analysis of all haplotypes, including the outgroups (Laurelia sempervirens, Nemuaron vieillardii and Daphnandra tenuipes), was undertaken using exhaustive searches for most-parsimonious trees with PAUP* version 4·0b10 (Swofford, 2000). The non-overlapping forward and reverse sequences for the petN-psbM intergenic spacer were concatenated into a single alignment for these and all subsequent analyses. Indels were scored as binary characters, while SSR regions were scored as multistate characters with a maximum of four states. All characters, including indels and SSR regions, were treated as unordered and of equal weight. Eight bases of the trnT-trnL intron were excluded from the analysis as the alignment of this region was ambiguous. Branch support was assessed by bootstrap analysis (Felsenstein, 1985) with 1000 bootstrap replicates using the same search parameters as those in the parsimony analysis. The K1-matK1 and petN-psbM sequences were of poor quality for the outgroup D. tenuipes and were scored as missing data.
DNA sequence based analyses and comparison with other species
Nucleotide diversity (π; Nei, 1987), the average number of nucleotide differences per site (excluding indels) between any two randomly chosen DNA sequences, of A. moschatum was calculated with the program DnaSP version 5·10 (Librado and Rozas, 2009) separately for each cpDNA fragment alignment (petN-psbM, trnL-trnT, trnL intron, K1-matK1 and matK6-K2) and for a concatenated alignment of all fragments. For comparison, the DNA sequence datasets of Nothofagus cunninghamii (213 samples; 2164 bp; trnS-trnfM, trnL-trnF, psbM-trnD, petN-psbM) from Worth et al. (2009) and Tasmannia lanceolata (244 samples; 3206 bp; petN-psbM, psbM-trnD, trnL-trnF, trnL intron, K1-matK1 and matK6-K2) from Worth et al. (2010) were also analysed. Alignments were constructed with MEGA 4·0 implementing the ClustalW program (Tamura et al., 2007). In addition, indel diversity per site [π(I)] was analysed for each species, with the multi-allelic option selected, using DnaSP version 5·10 (Librado and Rozas, 2009).
The distribution of pairwise mismatches (i.e. SNP differences between sequences, excluding indels) between cpDNA haplotypes was investigated using DnaSP version 5·10 (Librado and Rozas, 2009). For each species, this was undertaken using four different datasets; the whole species sampled range, and samples from three major regions, western Tasmania, eastern Tasmania and mainland Australia. Eastern Tasmania was defined as the region east of a line passing through the centre of the Midlands (Fig. 1) and extending into the Bass Strait, while western Tasmania was defined as being west of this line. This separation is based on the presence of the apparently significant biogeographic barrier formed by the broad, low, dry valley of the Midlands, and is consistent with phylogeographic structuring in N. cunninghamii and T. lanceolata (Worth et al., 2009, 2010).
Estimates of cpDNA substitution rates
The nucleotide substitution rates per million years (SSMY) were estimated for concatenated alignments by calculating the mean number of nucleotide substitutions per site (Nei, 1987) with Jukes and Cantor (1969) correction between all ingroup sequences and outgroups using DnaSP version 5·10 (Librado and Rozas, 2009). The outgroups were Laurelia sempervirens and Nemuaron viellardii for A. moschatum, Drimys winteri and Pseudowintera colorata for T. lanceolata, and N. glauca and N. menziesii for N. cunninghamii, with sequence data for Tasmannia and Nothofagus from Worth et al. (2010) and Worth et al. (2009), respectively. Estimated dates of divergence (mya) of ingroups and outgroups taxa were obtained from fossil calibrated molecular based phylogenies from Renner (1999) and Renner et al. (2000) for Atherospermataceae, Marquínez et al. (2009) for Winteraceae and, for Nothofagus subgenus Lophozonia, Cook and Crisp (2005) and Knapp et al. (2005).
RESULTS
Chloroplast variation
Five SNPs (two transitions and three transversions) and three 1-bp indels (for description of A. moschatum chloroplast polymorphisms see Supplementary Data Table S3) were observed within the 3294 bp of aligned sequence obtained for all 155 A. moschatum samples. All indels were associated with A or T mononucleotide repeats (SSRs) over 10 bp in length. Aligned sequence lengths and GenBank accession numbers for each fragment used in the preliminary screening and full sample set are shown in Supplementary Data Table S4.
Phylogenetic relationships
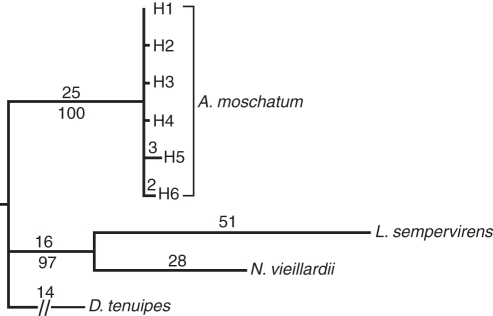
The eight polymorphisms observed in A. moschatum defined six haplotypes which formed a haplotype network without loops (Fig. 2). Parsimony analysis (using the outgroups and all A. moschatum haplotypes) resulted in four trees (36 characters parsimony informative; length = 142; consistency index = 0·99; retention index = 0·97) (Fig. 3). Bootstrap support (BP) was high for the monophyly of A. moschatum (BP = 100 %; Fig. 3) and for a clade containing L. sempervirens and N. vieillardii (BP = 97 %).
Fig. 3.
One of four most-parsimonious trees obtained from chloroplast sequence characters in Atherosperma moschatum and outgroups. This tree is identical to the strict consensus. Branch lengths (the inferred number of single base pair substitutions and indels on a branch) greater than one are indicated above branches while bootstrap values above 50 % are shown below branches. Names of A. moschatum haplotypes are the same as in Fig. 2. The break in the line for D. tenuipes indicates that data were missing.
Geographic distribution of haplotypes
The six haplotypes observed in A. moschatum showed geographic structure and generally occupied separate regions across the species latitudinal range. In Tasmania, three haplotypes (H1, H2 and H4) were observed among the 106 samples. Haplotype H1 (55 % of all A. moschatum samples) was widespread including the King and Flinders Islands in Bass Strait, but was not found among the 49 samples from mainland Australia (Fig. 2). Haplotype H2 was rare (2·6 % of all samples) and was observed in north-east Tasmania (n = 2), north-central Tasmania (Liffey Falls; n = 1) and south-eastern Tasmania (Tasman Peninsula; n = 1). Haplotype H4 (10·3 % of all samples) was observed only in central and south-west Tasmania, Bruny Island and the Tasman Peninsula. This latter site contained all three Tasmanian haplotypes (Fig. 2), making it the most diverse region sampled. Haplotype H3 (18·7 % of all samples) was the only haplotype observed in Victoria and in south-eastern NSW at Brown Mountain and the Geehi area, Kosciusko National Park (Fig. 2). Samples of the northern subspecies, A. moschatum subsp. integrifolium, harboured two different haplotypes differing by five polymorphisms. All three samples from the isolated population at Monga National Park possessed haplotype H5 (1·9 % of all samples), which was not found anywhere else. The 17 samples from three isolated populations in the northern parts of the range shared haplotype H6 (11 % of all samples). Overall, four of the haplotypes (H1, H2, H3 and H6) transgressed some major areas of unsuitable habitat, the Midlands of Tasmania for H1 and H2, parts of Bass Strait for H1, the rain shadow of the Bairnsdale-Omeo corridor (Howard and Ashton, 1973) for H3 and the Hunter Valley in NSW for H6.
DNA sequence-based analyses and comparison with other species
The mean nucleotide diversity for all 155 samples of A. moschatum was 0·00021, which is 4·8 and 3·5 times lower than that observed in the co-occurring species, Nothofagus cunninghamii and Tasmannia lanceolata respectively (Table 1). Nucleotide diversities differed between individual fragments for each species (Table 1). Indel diversity per site was also lower in A. moschatum (0·00012) than in either N. cunninghamii (0·00019) or T. lanceolata (0·00031).
Table 1.
The nucleotide diversity (π × 10−3) estimated for Nothofagus cunninghamii (213 samples), Tasmannia lanceolata (244 samples) and Atherosperma moschatum (155 samples) for each fragment and the whole alignments
| N. cunninghamii | T. lanceolata | A. moschatum | |
|---|---|---|---|
| trnS-trnfM | 2·16 ± 0·14 | – | – |
| rps16 intron | 0·05 ± 0·04 | – | – |
| trnL-trnF | 1·66 ± 0·26 | 0·35 ± 0·11 | * |
| psbM-trnD | 1·09 ± 0·18 | 2·14 ± 0·21 | (0·00 ± 0·00) |
| petN-psbM | 0·14 ± 0·05 | 0·27 ± 0·08 | 0·23 ± 0·05 |
| K1-matK1 | – | 0·44 ± 0·07 | 0·40 ± 0·07 |
| matK6-K2 | – | 0·38 ± 0·05 | 0·24 ± 0·05 |
| trnL intron | – | 0·18 ± 0·05 | 0·00 ± 0·00 |
| trnT-trnL | – | – | 0·08 ± 0·05 |
| atpB-rbcL | – | – | (0·00 ± 0·00) |
| Whole alignment | 1·01 ± 0·1 | 0·73 ± 0·06 | 0·21 ± 0·03 |
The highest values for each fragment are indicated in bold.
A dash denotes fragments that were not studied for the species and an asterisk denotes a fragment that did not amplify.
Nucleotide diversities in parenthesis were calculated with only 24 samples.
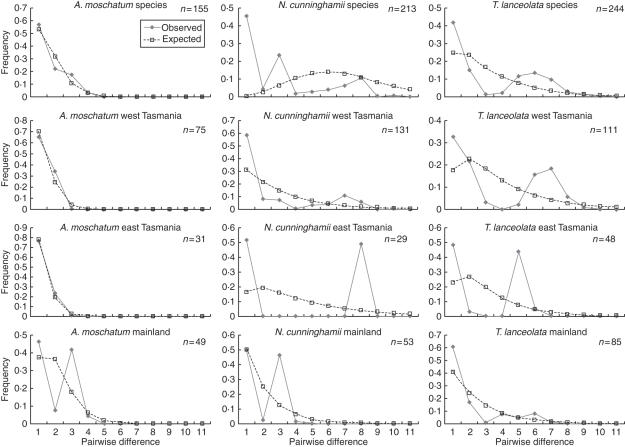
The distributions of pairwise mismatches between cpDNA haplotypes were strongly multimodal in N. cunninghamii and T. lanceolata at the species level and when analysed for different regions (Fig. 4). In contrast, for A. moschatum, the mismatch distributions were unimodal, apart from the mainland where the most diverged haplotypes in A. moschatum were observed in the most northern part of the species' mainland range.
Fig. 4.
Distributions of pairwise mismatches between cpDNA haplotypes for the species (Atherosperma moschatum, Nothofagus cunninghamii and Tasmannia lanceolata) and populations in three regions (western Tasmania, eastern Tasmania and south-east mainland Australia). Expected frequencies of pairwise differences in a growth/decline population model are indicated by broken lines. Observed frequencies of pairwise differences between haplotypes are indicated by continuous lines. Note the marked bi- or trimodality in all regions for both N. cunninghamii and T. lanceolata, but mostly unimodal distributions for A. moschatum.
Comparative mutation rates
Rates of SSMY were between 1·79 and 3·34 × 10−4 for the branch leading to A. moschatum when estimated using the calibration with Laurelia sempervirens, while they were between 1·64 and 5·68 × 10−4 when using Nemauron veillardii for calibration (see Supplementary Data Table S5). Apart from the higher rates when using the youngest divergence times of A. moschatum and outgroup species, these values were similar to the cpDNA substitution rate of rbcL inferred for the family Atherospermataceae of 1·4–2·4 × 10−4 SSMY by Renner et al. (2000).
DISCUSSION
Low chloroplast variation
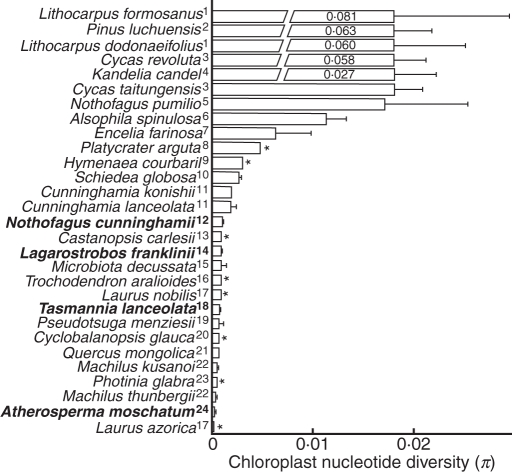
The chloroplast variation observed in A. moschatum appears to be unusually low, with lower estimated nucleotide diversity than 27 of 28 other woody plant species for which values were available (Fig. 5). The only lower estimate was based on a comparatively smaller sample size in a rare and mostly insular tree species, Laurus azorica (Rodriguez-Sanchez et al., 2009). Although direct comparisons of cpDNA diversity with other plant species can be affected by the choice of cpDNA fragments and the intensity and coverage of sampling, the value for A. moschatum is so low that, in conjunction with the widespread sampling of its range, it appears unlikely to be due to such effects. We argue that the paucity of variation is likely to be due to a reduction of genetic diversity in the species (i.e. via bottlenecks) rather than a recent origin of the species or low rates of molecular evolution.
Fig. 5.
Chloroplast nucleotide diversity (π; Nei, 1987) ± s.e. for 29 woody plant species, including Atherosperma moschatum. All Australian cool temperate rainforest species are shown in bold. High values have been truncated with actual values shown on the bar. No standard errors were available for species indicated with an asterisk. Only values calculated from species with more than ten samples were included. Sources: 1, Chiang et al. (2004); 2, Chiang et al. (2006); 3, Chiang et al. (2009); 4, Chiang et al. (2001); 5, Mathiasen and Premoli (2009); 6, Su et al. (2005); 7, Fehlberg and Ranker (2009); 8, Qiu et al. (2009); 9, Ramos et al. (2009); 10, Wallace et al. (2009); 11, Hwang et al. (2003); 12, Worth et al. (2009); 13, Cheng et al. (2005); 14, Clark and Carbone (2008); 15, Artyukova et al. (2009); 16, Huang et al. (2004); 17, Rodriguez-Sanchez et al. (2009); 18, Worth et al. (2010); 19, Gugger et al. (2010); 20, Huang et al. (2002); 21, Okaura et al. (2007); 22, Wu et al. (2006); 23, Aoki et al. (2006); 24, this study. Full details are given in Supplementary Data Table S6 (available online).
This low genetic diversity is unlikely to be the result of recent evolution or arrival in south-eastern Australia. This is because the genus is monotypic and endemic to the region, and molecular dating based on a phylogeny encompassing all genera in the family and the sequencing of 4203 bp of the chloroplast genome provides an age of 69·5 million years for the genus (Renner, 2004). Furthermore, there is macrofossil evidence (a leaf fossil that is indistinguishable from leaves of the extant species) for the presence of the species in western Tasmania in the Early Pleistocene, approx. 1 million years ago (Hill and Macphail, 1985).
The possibility that the low diversity of A. moschatum can be explained by low cpDNA substitution rates can be discounted for this species. It is plausible that rates of molecular evolution may have been relatively low in A. moschatum because it has the ability to regenerate asexually from basal sprouts, possibly extending the lifespan of individuals (Hickey et al., 1982; Read and Hill, 1988b), a trait that can be associated with low rates of mutation in angiosperms (Smith and Donoghue, 2008). However, the cpDNA substitution rates estimated for A. moschatum (between 1·64–5·68× 10−4 SSMY) were similar or higher in comparison to values estimated for T. lanceolata (1·28–1·51 × 10−4 SSMY) and N. cunninghamii (1·23–2·6 × 10−4 SSMY) in this study and by Albert et al. (1994) for both Winteraceae (0·98 to 1·47 × 10−4 SSMY) and Nothofagaceae (2·10 × 10−4 SSMY). Thus, although there is considerable uncertainty in substitution rate estimates (Renner et al., 2000; Kay et al., 2006) there is no reason to suppose that A. moschatum has slower molecular evolution of the chloroplast than co-occurring species with higher levels of chloroplast diversity.
One possible explanation for the low chloroplast diversity of A. moschatum would be major reductions in population size (i.e. bottlenecks) in the past due to cold intolerance. This hypothesis is consistent with marked ecological and physiological differences between A. moschatum and the more genetically diverse species N. cunninghamii and T. lanceolata. Unlike A. moschatum which rarely occurs near or above the tree line (Hill et al., 1988), both N. cunninghamii and T. lanceolata form large populations well above the tree-line (Macphail, 1975). Experimental evidence suggests that A. moschatum is the least cold tolerant of the tree species in cool temperate rainforest (Read and Hill, 1989; Read and Busby, 1990; Feild and Brodribb, 2001). For example, after a single freeze–thaw cycle, A. moschatum was shown to be the most vulnerable of five other Tasmanian rainforest woody species to loss of hydraulic conductivity (Feild and Brodribb, 2001). In addition, A. moschatum had the lowest frost resistance and lowest photosynthetic tolerance to low temperatures of all other Australian cool temperate trees studied (Read and Hill, 1989; Read and Busby, 1990).
During the LGM, treeless conditions are thought to have extended as far north as Barrington Tops (Sweller and Martin, 2001). Stands of tall forest, like those in the present day, would have been highly restricted in the LGM due to lower rainfall and a depressed tree-line, with the tree-line being near current sea-level in Tasmania (Colhoun, 1985). Based on evidence of the extent of glaciation, Early Pleistocene glacials may have been colder than the LGM (Kiernan, 1990; Colhoun et al., 1996). Thus, the habitat for A. moschatum during glacials may have been considerably more restricted than those of N. cunninghamii and T. lanceolata resulting in very small populations of A. moschatum, thereby reducing existing variation by genetic drift (i.e. the bottleneck effect). Moreover, such bottlenecks may have occurred repeatedly over the Pleistocene glacial/interglacial cycles.
Repeated bottlenecks, dispersal and multiple refugia
The star-like chloroplast network observed in A. moschatum implies that the species has undergone a major bottleneck, or the cumulative effect of multiple bottlenecks, that reduced the chloroplast diversity within the species to a single haplotype in the past. An alternative explanation is that this major loss of diversity in A. moschatum may have involved a selective sweep, although this is difficult to detect with chloroplast data alone. This haplotype was most likely to have been haplotype H1 or an extinct ancestor, the zero branch length leading to haplotype H1 on the phylogram (Fig. 3) suggesting that haplotype H1 is plesiomorphic, retaining ancestral states to all other haplotypes observed in A. moschatum. Any major bottleneck and subsequent recovery leading to the occupation of its current widespread range are unlikely to have occurred in the recent past (i.e. since the LGM) as this scenario would require rapid evolution of five haplotypes during range expansion, two of which are characterized by multiple differences from the ancestral hapotype H1. In fact, the modern geographic structuring of haplotypes is likely to have developed over multiple glacial periods involving establishment of new populations via rare dispersal events and glacial survival and bottlenecks (i.e. refugia) within multiple regions. The clearest evidence for the effect of glacial refugia comes from Victoria and NSW, where three regions contain a single, endemic-derived haplotype. Apart from the possibility of selection, this chloroplast divergence is best explained by bottlenecking events subsequent to the major event described above, because it involves either the in situ loss of ancestral haplotypes or dispersal of the derived haplotype to the new region and extinction of that same haplotype in its ancestral area.
On the mainland the chloroplast evidence supports survival within at least one glacial refugia in each of three regions: near Monga in south-eastern NSW; the Blue Mountains/Barrington Tops region NSW; and within the current range of haplotype H3 in Victoria/far south-eastern NSW. The chloroplast evidence for the location of glacial refugia for A. moschatum in Tasmania is less clear. Haplotype H4 provides evidence for survival through the last glacial in at least one area in Tasmania, probably in western Tasmania where LGM fossil pollen of the species, albeit at very low levels, has been found (Macphail and Colhoun, 1985; Colhoun et al., 1999; Hopf et al., 2000). In addition, it is possible that the Tasman Peninsula, where all three Tasmanian haplotypes were found, represents an additional LGM refugia, an assertion that is supported by genetic evidence for two wet forest Eucalyptus species (Bloomfield et al., 2011; Nevill et al., 2010). However, the chloroplast pattern in this region could equally be explained by Holocene admixture, particularly given the absence of any fossil pollen record from this region. However, the sharing of single haplotypes across large geographic areas, particularly for H1 in Tasmania, H3 across the whole species range in Victoria and south-eastern NSW and H6 on both sides of the Hunter Valley, a dryland barrier for wet forest species situated between the Blue Mountains and Barrington Tops regions, limits the ability to locate precisely glacial refugia for the species. This is in contrast to N. cunninghamii and, in particular, T. lanceolata where high haplotype diversity and endemicity allowed comparatively fine-scale resolution of the location of refugia (Worth et al., 2009, 2010).
Some hypothesis can also be made of the dispersal of the species. On mainland south-eastern Australia where populations are generally small and geographically isolated, all populations sampled contained a single haplotype. This contrasts with previous chloroplast phylogeographic studies of some trees with plumose wind-dispersed seeds. For example, admixture of haplotypes was observed across the range (98 samples) of Populus tremula in Italy (Salvini et al., 2001). A similar finding was observed in Salix caprea across Europe (Palme et al., 2003). Despite having a similar dispersal mechanism, A. moschatum may differ from these aforementioned tree species due to its non-pioneer status and generally poor recruitment (Hickey et al., 1982; Read, 1985; Neyland, 1991), resulting in the relatively rare successful establishment of viable populations in either new habitat or already occupied areas. However, the constraints on dispersal may be much lower than in the co-occurring species, Nothofagus cunninghamii and Tasmannia lanceolata, which show strong, relatively small-scale geographic patterning of chloroplast haplotypes (Worth et al., 2009, 2010). In mainland populations of A. moschatum, two additional factors are likely to have contributed to the lack of admixture of haplotypes – the fact that there are only three observed haplotypes in this region, and that mainland populations of the species are in most cases geographically separated by extensive inhospitable territory (>150 km in each case). Admixture of haplotypes may explain the co-occurrence of haplotypes in Tasmania, where the rare haplotype H2 occurs on both sides of the dry Midlands and all three Tasmanian haplotypes were found on the Tasman Peninsula. This discrepancy could be explained by a greater level of long-distance dispersal related to the greater propagule pressure of the species in Tasmania where the species is more common than on the mainland (Shapcott, 1994).
While it is plausible that the current distribution of A. moschatum has been impacted by extensive Holocene expansion within some regions, e.g. within Victoria, better understanding of the timing of any dispersal events across areas of unsuitable habitat is difficult and is likely to require the analysis of the nuclear markers such as nuclear microsatellites or sequence data. Nuclear markers may be particularly informative due to the fact that A. moschatum is insect pollinated and therefore likely to have restricted pollen dispersal (Shapcott, 1995). In addition, next generation sequencing technology could be used to simultaneously scan whole chloroplast genomes of multiple individuals (Parks et al., 2009) to investigate whether widespread haplotypes are in fact differentiated in other parts of the chloroplast genome not investigated in this study.
Conclusions
Although A. moschatum is the most widespread of all cool temperate rainforest woody plants in Australia, the species chloroplast diversity was the lowest of any cool temperate rainforest plant studied to date and among the lowest documented among woody plants. Similar estimated rates of chloroplast substitution to N. cunninghamii and T. lanceolata, species that both have higher chloroplast diversity, mismatch analysis and geographic patterning of haplotypes provide evidence that A. moschatum underwent successive bottlenecks reducing existing chloroplast variation during the Pleistocene. A severe bottleneck several glacials ago may have reduced chloroplast diversity to a single haplotype. Further bottlenecks occurred during subsequent glacials, including the LGM, resulting in the restriction of endemic haplotypes to four separate regions across its range, south-western Tasmania, Victoria/far south-eastern NSW, Monga, and the most northern populations in NSW. Dispersal to new areas of suitable habitat plausibly played a role in the Holocene within these regions; however, detailed information on the history of populations within these regions awaits further investigation.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank John and Noela Cross, Jon Marsden-Smedley, Peter Woodard and Dave Woods for help with sample collection; Ken Gillanders for access to his private collection; and the Department of Sustainability and Environment (Victoria), the Department of Primary Industries, Parks, Water and Environment (Tasmania) and the Department of Environment, Climate Change and Water (NSW) for collection permits. We also thank two anonymous reviewers for their comments which helped to improve this manuscript. This work was supported by a Discovery grant DP0557260 from the Australian Research Council awarded to René Vaillancourt.
LITERATURE CITED
- Albert VA, Backlund A, Bremer K, Chase MW, Manhart JR, Mishler BD, Nixon KC. Functional constraints and rbcL evidence for land plant phylogeny. Annals of the Missouri Botanical Garden. 1994;81:534–567. [Google Scholar]
- Aoki K, Matsumara T, Hattori T, Murukami N. Chloroplast DNA phylogeography of Photinia glabra (Rosaceae) in Japan. American Journal of Botany. 2006;93:1852–1858. doi: 10.3732/ajb.93.12.1852. [DOI] [PubMed] [Google Scholar]
- Artyukova EV, Kozyrenko MM, Gorovoy PG, Zhuravlev YN. Plastid DNA variation in highly fragmented populations of Microbiota decussata Kom. (Cupressaceae), an endemic to Sikhote Alin Mountains. Genetica. 2009;137:201–212. doi: 10.1007/s10709-009-9386-7. [DOI] [PubMed] [Google Scholar]
- Ashton D. The Big Ash forest, Wallaby Creek, Victoria: changes during one lifetime. Australian Journal of Botany. 2000;48:1–26. [Google Scholar]
- Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Blackman CJ, Brodribb TJ, Jordan GJ. Leaf hydraulic vulnerability is related to conduit dimensions and drought resistance across a diverse range of woody angiosperms. New Phytologist. 2010;188:1113–1123. doi: 10.1111/j.1469-8137.2010.03439.x. [DOI] [PubMed] [Google Scholar]
- Bloomfield JA, Nevill P, Potts BM, Vaillancourt RE, Steane DA. Molecular genetic variation in a widespread forest tree species Eucalyptus obliqua (Myrtaceae) on the island of Tasmania. Australian Journal of Botany. 2011;59:226–237. [Google Scholar]
- Brophy JJ, Buchanan AM, Copeland LM, Dimitriadis E, Goldsack RJ, Hibbert DB. Differentiation between the two subspecies of Atherosperma moschatum Labill. (Atherospermataceae) from their leaf oils. Biochemical Systematics and Ecology. 2009;37:479–483. [Google Scholar]
- Carpenter RJ, Jordan GJ. Early Tertiary macrofossils of Proteaceae from Tasmania. Australian Systematic Botany. 1997;10:533–563. [Google Scholar]
- Cheng Y, Hwang S, Lin T. Potential refugia in Taiwan revealed by the phylogeographical study of Castanopsis carlesii Hayata (Fagaceae) Molecular Ecology. 2005;14:2075–2085. doi: 10.1111/j.1365-294X.2005.02567.x. [DOI] [PubMed] [Google Scholar]
- Chiang T, Hung K, Hsu T, Wu W. Lineage sorting and phylogeography in Lithocarpus formosanus and L. dodonaeifolius (Fagaceae) from Taiwan. Annals of the Missouri Botanical Gardens. 2004;91:207–222. [Google Scholar]
- Chiang TY, Chiang YC, Chen YJ, et al. Phylogeography of Kandelia candel in East Asiatic mangroves based on nucleotide variation of chloroplast and mitochondrial DNAs. Molecular Ecology. 2001;10:2697–2710. doi: 10.1046/j.0962-1083.2001.01399.x. [DOI] [PubMed] [Google Scholar]
- Chiang Y, Hung K, Schaal B, Ge X, Hsu T, Chiang T. Contrasting phylogeographical patterns between mainland and island taxa of the Pinus luchuensis complex. Molecular Ecology. 2006;15:765–779. doi: 10.1111/j.1365-294X.2005.02833.x. [DOI] [PubMed] [Google Scholar]
- Chiang YC, Hung KH, Moore SJ, et al. Paraphyly of organelle DNAs in Cycas Sect. Asiorientales due to ancient ancestral polymorphisms. BMC Evolutionary Biology. 2009;9:161. doi: 10.1186/1471-2148-9-161. doi:10.1186/1471-2148-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark C, Carbone I. Chloroplast DNA phylogeography in long-lived Huon pine, a Tasmanian rain forest conifer. Canadian Journal of Botany. 2008;38:1576–1589. [Google Scholar]
- Colhoun EA. Pre-last glaciation maximum vegetation history at Henty Bridge, western Tasmania. New Phytologist. 1985;100:681–690. [Google Scholar]
- Colhoun EA, Hannah D, Kiernan K. Late Wisconsin glaciation of Tasmania. Papers and Proceedings of the Royal Society of Tasmania. 1996;130:33–45. [Google Scholar]
- Colhoun EA, Pola JS, Barton CE, Heijnis H. Late Pleistocene vegetation and climate history of Lake Selina, western Tasmania. Quaternary International. 1999;57–58:5–23. [Google Scholar]
- Cook LG, Crisp MD. Not so ancient: the extant crown group of Nothofagus represents a post-Gondwanan radiation. Proceedings of the Royal Society B: Biological Sciences. 2005;272:2535–2544. doi: 10.1098/rspb.2005.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis W. The student's flora of Tasmania. Angiospermae: Plumbaginaceae to Salicaceae. Hobart, Australia: St. David's Park Publishing; 1993. [Google Scholar]
- Coyne P. Protecting the natural treasures of the Australian Alps. 2001 Natural Heritage Working Group of the Australian Alps Liaison Committee. [Google Scholar]
- Davis MB. Pleistocene biogeography of temperate deciduous forests. Geoscience and Man. 1976;13:13–26. [Google Scholar]
- Feild TS, Brodribb T. Stem water transport and freeze–thaw xylem embolism in conifers and angiosperms in a Tasmanian treeline heath. Oecologia. 2001;127:314–320. doi: 10.1007/s004420000603. [DOI] [PubMed] [Google Scholar]
- Fehlberg S, Ranker T. Evolutionary history and phylogeography of Encelia farinosa (Asteraceae) from the Sonoran, Mojave, and Peninsular Deserts. Molecular Phylogenetics and Evolution. 2009;50:326–335. doi: 10.1016/j.ympev.2008.11.011. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Floyd AG. Rainforest trees of mainland southeastern Australia. Melbourne: Inkata Press; 1989. [Google Scholar]
- Floyd AG. Australian rainforests in New South Wales. Chipping Norton, NSW: Surrey Beatty and Sons; 1990. [Google Scholar]
- Foreman DB, Whiffen T. Atherospermataceae. In: Wilson AG, editor. Flora of Australia. Melbourne: ABRS; 2007. [Google Scholar]
- Gellie NJH. Native vegetation of the southern forests: south-east highlands, Australian Alps, south-west slopes, and SE corner bioregions. Cunninghamia. 2005;9:219–252. [Google Scholar]
- Gilbert JM. Forest succession in the Florentine Valley, Tasmania. Papers & Proceedings of the Royal Society of Tasmania. 1959;93:129–151. [Google Scholar]
- Gugger P, Sugita S, Cavender-Bares J. Phylogeography of Douglas-fir based on mitochondrial and chloroplast DNA sequences: testing hypotheses from the fossil record. Molecular Ecology. 2010;19:1877–1897. doi: 10.1111/j.1365-294X.2010.04622.x. [DOI] [PubMed] [Google Scholar]
- Hickey JE, Blakesley AJ, Turner B. Seedfall and germination of Nothofagus cunninghamii (Hook.) Oerst., Eucryphia lucida (Labill.) Baill. and Atherosperma moschatum Labill.: implications for regeneration practice. Australian Forest Research. 1982;13:21–28. [Google Scholar]
- Hill RS. Origins of the southeastern Australian vegetation. Philosophical Transactions of the Royal Society of London Series B. 2004;359:1537–1549. doi: 10.1098/rstb.2004.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RS, Macphail MK. A fossil flora from rafted Plio-Pleistocene mudstones at Regatta Point, Tasmania. Australian Journal of Botany. 1985;33:497–517. [Google Scholar]
- Hill RS, Read J, Busby JR. The temperature-dependence of photosynthesis of some Australian temperate rainforest trees and its biogeographical significance. Journal of Biogeography. 1988;15:431–449. [Google Scholar]
- Hopf FVL, Colhoun EA, Barton CE. Late-glacial and Holocene record of vegetation and climate from Cynthia Bay, Lake St Clair, Tasmania. Journal of Quaternary Science. 2000;15:725–732. [Google Scholar]
- Howard T, Ashton D. The distribution of Nothofagus cunninghamii rainforest. Proceeding of the Royal Society of Victoria. 1973;85:47–75. [Google Scholar]
- Huang S, Hwang S, Lin T. Spatial pattern of chloroplast DNA variation of Cyclobalanopsis glauca in Taiwan and East Asia. Molecular Ecology. 2002;11:2349–2358. doi: 10.1046/j.1365-294x.2002.01624.x. [DOI] [PubMed] [Google Scholar]
- Huang S, Hwang S, Wang J, Lin T. Phylogeography of Trochodendron aralioides (Trochodendraceae) in Taiwan and its adjacent areas. Journal of Biogeography. 2004;31:1251–1259. [Google Scholar]
- Huntley B, Birks H. An atlas of past and present pollen maps for Europe: 0–13000 years ago. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Huntley B, Webb T. Migration: species' response to climatic variations caused by changes in the earth's orbit. Journal of Biogeography. 1989;16:5–19. [Google Scholar]
- Hwang S-H, Lin T-P, Ma C-S, Lin C-L, Chung J-D, Yang J-C. Postglacial population growth of Cunninghamia konishii (Cupressaceae) inferred from phylogeographical and mismatch analysis of chloroplast DNA variation. Molecular Ecology. 2003;12:2689–2695. doi: 10.1046/j.1365-294x.2003.01935.x. [DOI] [PubMed] [Google Scholar]
- Jordan GJ. Evidence of Pleistocene plant extinction and diversity from Regatta Point, western Tasmania, Australia. Botanical Journal of the Linnean Society. 1997;123:45–71. [Google Scholar]
- Jukes T, Cantor C. Evolution of protein molecules. In: Munro MN, editor. Mammalian protein metabolism. New York, NY: Academic Press; 1969. [Google Scholar]
- Kay KM, Whittall JB, Hodges SA. A survey of nuclear ribosomal internal transcribed spacer substitution rates across angiosperms: an approximate molecular clock with life history effects. BMC Evolutionary Biology. 2006;6:36. doi: 10.1186/1471-2148-6-36. doi:10.1186/1471-2148-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kershaw AP, McKenzie GM, Porch N, et al. A high-resolution record of vegetation and climate through the last glacial cycle from Caledonia Fen, southeastern highlands of Australia. Journal of Quaternary Science. 2007;22:481–500. [Google Scholar]
- Kiernan K. The extent of Late Cenozoic glaciation in the Central Highlands of Tasmania, Australia. Arctic & Alpine Research. 1990;22:341–354. [Google Scholar]
- Kirkpatrick J. A transect study of forests and woodlands on dolerite in the Eastern Tiers, Tasmania. Vegetatio. 1981;44:155–163. [Google Scholar]
- Knapp M, Stockler K, Havell D, Delsuc F, Sebastiani F, Lockhart P. Relaxed molecular clock provides evidence for long-distance dispersal of Nothofagus (southern beech) PLoS Biology. 2005;3:e14. doi: 10.1371/journal.pbio.0030014. doi:10.1371/journal.pbio.0030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Lunt I. Atherosperma moschatum (southern sassafras) in the Otway Ranges: a victim of European landuse? Victorian Naturalist. 1992;109:85–86. [Google Scholar]
- McKenzie GM, Kershaw AP. A vegetation history and quantitative estimate of Holocene climate from Chapple Vale, in the Otway region of Victoria, Australia. Australian Journal of Botany. 1997;45:565–581. [Google Scholar]
- Macphail MK. Late Pleistocene environments in Tasmania. Search. 1975;6:295–300. [Google Scholar]
- Macphail MK, Colhoun EA. Late last glacial vegetation, climates and fire activity in southwest Tasmania. Search. 1985;16:43–45. [Google Scholar]
- Marquínez X, Lohmann LG, Faria Salatino ML, Salatino A, González F. Generic relationships and dating of lineages in Winteraceae based on nuclear (ITS) and plastid (rpS16 and psbA-trnH) sequence data. Molecular Phylogenetics and Evolution. 2009;53:435–449. doi: 10.1016/j.ympev.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Mathiasen P, Premoli AC. Out in the cold: genetic variation of Nothofagus pumilio (Nothofagaceae) provides evidence for latitudinally distinct evolutionary histories in austral South America. Molecular Ecology. 2009;19:371–385. doi: 10.1111/j.1365-294X.2009.04456.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York, NY: Columbia University Press; 1987. [Google Scholar]
- Nevill PG, Bossinger G, Ades PK. Phylogeography of the world's tallest angiosperm, Eucalyptus regnans: evidence for multiple isolated Quaternary refugia. Journal of Biogeography. 2010;37:179–192. [Google Scholar]
- Neyland M. Relict rainforest in eastern Tasmania. Tasmanian NRCP Technical Report No. 6. Parks and Wildlife Heritage. 1991 Canberra, Hobart and Department of Arts, Sport, the Environment, Tourism and Territories. [Google Scholar]
- Neyland M, Brown M. Rainforest in eastern Tasmania: floristics and conservation. Papers and Proceedings of the Royal Society of Tasmania. 1993;127:23–32. [Google Scholar]
- Okaura T, Quang N, Ubukata M, Harada K. Phylogeographic structure and late Quaternary population history of the Japanese oak Quercus mongolica var. crispula and related species revealed by chloroplast DNA variation. Genes and Genetic Systems. 2007;82:465–477. doi: 10.1266/ggs.82.465. [DOI] [PubMed] [Google Scholar]
- Palme AE, Semerikov V, Lascoux M. Absence of geographical structure of chloroplast DNA variation in sallow, Salix caprea L. Heredity. 2003;91:465–474. doi: 10.1038/sj.hdy.6800307. [DOI] [PubMed] [Google Scholar]
- Parks M, Cronn R, Liston A. Increasing phylogenetic resolution at low taxonomic levels using massively parallel sequencing of chloroplast genomes. BMC Biology. 2009;7:84. doi: 10.1186/1741-7007-7-84. doi:10.1186/1741-7007-7-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu YX, Qi XS, Jin XF, et al. Population genetic structure, phylogeography, and demographic history of Platycrater arguta (Hydrangeaceae) endemic to East China and South Japan, inferred from chloroplast DNA sequence variation. Taxon. 2009;58:1226–1241. [Google Scholar]
- Ramos ACS, De Lemos-Filho JP, Lovato MB. Phylogeographical structure of the neotropical forest tree Hymenaea courbaril (Leguminosae: Caesalpinioideae) and its relationship with the vicariant Hymenaea stigonocarpa from Cerrado. Journal of Heredity. 2009;100:206–216. doi: 10.1093/jhered/esn092. [DOI] [PubMed] [Google Scholar]
- Read J. Photosynthetic and growth responses to different light regimes of the major canopy species of Tasmanian cool temperate rainforest. Australian Journal of Ecology. 1985;10:327–334. [Google Scholar]
- Read J, Busby JR. Comparative responses to temperature of the major canopy species of Tasmanian cool temperate rainforest and their ecological significance. II. Net photosynthesis and climate analysis. Australian Journal of Botany. 1990;38:185–205. [Google Scholar]
- Read J, Hill RS. Comparative responses to temperature of the major canopy species of Tasmanian cool temperate rainforest and their ecological significance. I. Foliar frost resistance. Australian Journal of Botany. 1988a;36:131–143. [Google Scholar]
- Read J, Hill RS. The dynamics of some rainforest associations in Tasmania. Journal of Ecology. 1988b;76:558–584. [Google Scholar]
- Read J, Hill RS. The response of some Australian temperate rain forest tree species to freezing temperatures and its biogeographical significance. Journal of Biogeography. 1989;16:21–27. [Google Scholar]
- Renner S. Circumscription and phylogeny of the Laurales: evidence from molecular and morphological data. American Journal of Botany. 1999;86:1301–1315. [PubMed] [Google Scholar]
- Renner SS. Variation in diversity among Laurales, Early Cretaceous to Present. Biologiske Skrifter. 2004;55:441–458. [Google Scholar]
- Renner S, Foreman D, Murray D. Timing transantarctic disjunctions in the Atherospermataceae (Laurales): evidence from coding and noncoding chloroplast sequences. Systematic Biology. 2000;49:579–591. doi: 10.1080/10635159950127402. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Sanchez F, Guzmin B, Valido A, Vargas P, Arroyo J. Late Neogene history of the laurel tree (Laurus L., Lauraceae) based on phylogeographical analyses of Mediterranean and Macaronesian populations. Journal of Biogeography. 2009;36:1270–1281. [Google Scholar]
- Salvini D, Anzidei M, Fineschi S, Emilia-Malvolti M, Taurchini D, Vendramin GG. Low genetic differentiation among Italian populations of Populus tremula L. (Salicaceae) estimated using chloroplast PCR-RFLP and microsatellite markers. Forest Genetics. 2001;8:81–87. [Google Scholar]
- Schodde R. A monograph of the family Atherospermataceae. 1969 PhD Thesis, University of Adelaide. [Google Scholar]
- Shapcott A. Genetic and ecological variation in Atherosperma moschatum and the implications for conservation of its biodiversity. Australian Journal of Botany. 1994;42:663–686. [Google Scholar]
- Shapcott A. The spatial genetic structure in natural populations of the Australian temperate rainforest tree Atherosperma moschatum (Labill.) (Monimiaceae) Heredity. 1995;74:28–38. [Google Scholar]
- Smith AS, Donoghue MJ. Rates of molecular evolution are linked to life history in flowering plants. Science. 2008;322:86–89. doi: 10.1126/science.1163197. [DOI] [PubMed] [Google Scholar]
- Sniderman JMK, Jordan GJ. Extent and timing of floristic exchange between Australian and Asian rain forests. Journal of Biogeography. 2011;38:1445–1455. [Google Scholar]
- Su Y, Wang T, Zheng B, et al. Genetic differentiation of relictual populations of Alsophila spinulosa in southern China inferred from cpDNA trnL-F noncoding sequences. Molecular Phylogenetics and Evolution. 2005;34:323–333. doi: 10.1016/j.ympev.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Sweller S, Martin H. A 40,000 year vegetation history and climatic interpretations of Burraga Swamp, Barrington Tops, New South Wales. Quaternary International. 2001;83–85:233–244. [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods), version 4·0b10. Sunderland, MA: Sinauer; 2000. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4·0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Wallace LE, Weller SG, Wagner WL, Sakai AK, Nepokroeff M. Phylogeographic patterns and demographic history of Schiedea globosa (Caryophyllaceae) on the Hawaiian Islands. American Journal of Botany. 2009;96:958–967. doi: 10.3732/ajb.0800243. [DOI] [PubMed] [Google Scholar]
- Worth J, Jordan G, McKinnon G, Vaillancourt R. The major Australian cool temperate rainforest tree Nothofagus cunninghamii withstood Pleistocene glacial aridity within multiple regions: evidence from the chloroplast. New Phytologist. 2009;182:519–532. doi: 10.1111/j.1469-8137.2008.02761.x. [DOI] [PubMed] [Google Scholar]
- Worth JRP, Jordan GJ, Marthick JR, McKinnon GE, Vaillancourt RE. Chloroplast evidence for geographic stasis of the Australian bird-dispersed shrub Tasmannia lanceolata (Winteraceae) Molecular Ecology. 2010;19:2949–2963. doi: 10.1111/j.1365-294X.2010.04725.x. [DOI] [PubMed] [Google Scholar]
- Wu S, Hwang C, Lin T, Chung J, Cheng Y, Hwang S. Contrasting phylogeographical patterns of two closely related species, Machilus thunbergii and Machilus kusanoi (Lauraceae), in Taiwan. Journal of Biogeography. 2006;33:936–947. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.