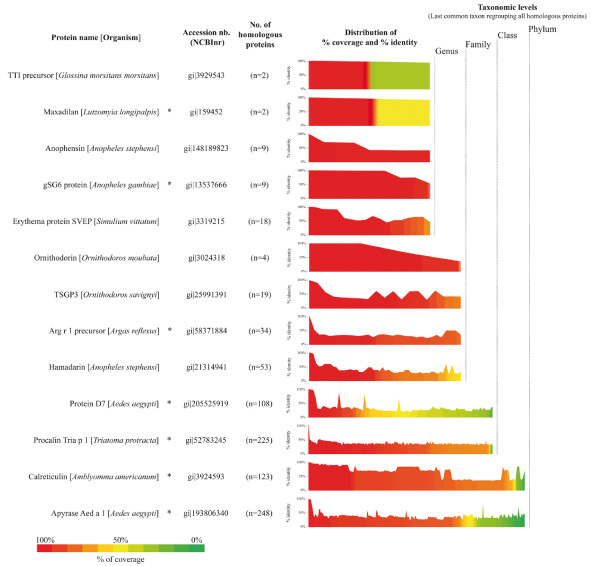
Figure 3.
Protein sequence diversity of haematophagous arthropods salivary proteins. Sequences of 13 salivary proteins of haematophagous arthropods described in the present review were submitted to BLAST analysis on the non-redundant protein database (NCBInr, NIH, Bethesda). The blastp program was used with default parameters excepting the following: search was done on the Arthropoda taxonomic level (taxid: 6656, Nov 15th, 2010, 12,289,957 sequences), the E-value threshold was changed to a setting of 1 in order to recover only hits with highest significance on the overall protein sequence and the hit-list size was set to 5000 proteins. The number of homologous proteins with a score above 40 and their respective percentage of coverage and identity were recovered for all query proteins and sorted according to their increasing percentage of coverage. The number of homologous proteins is indicated in brackets (this includes the query sequence) and the distributions of both the percentage of coverage (bar graph with a coloured scale) and the percentage of identity (line profile above each bar graph) are represented. Proteins are grouped according to the taxonomic level of the last common taxon regrouping their corresponding homologous proteins. For graphical convenience, subclass, class and infraclass as well as superfamily, family and subfamily taxonomic levels were grouped into class and family, respectively. Salivary proteins reported to be targeted by an immune response are indicated by an asterisk (*).