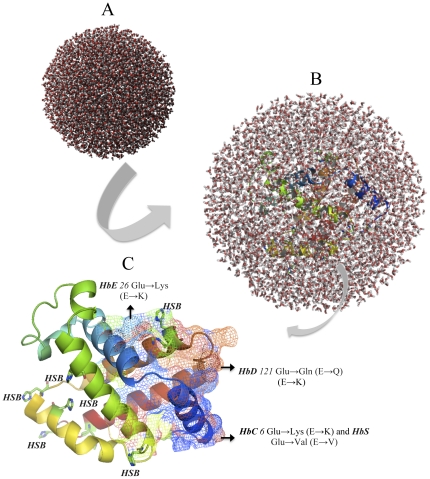
Figure 3. The ß-chain of the HHB protein's molecular dynamics (MD) simulation showing truncated octahedron boundary explicit water solvated and hydrogen atoms in color.
The MD simulations system used in calculations are; a) water box without the protein, b) water box surrounding the entire protein (middle) and c) tertiary structure of the wild type ß-chain of HHB protein showing disease causing mutations i.e. green HbE26 Glu→Lys (E→K), orange HbD121 Glu→Gln (E→Q), blue HbC6 Glu→Lys (E→K) and HbS6 Glu→Val (E→V) on 3 helixes using predict mesh modeling. The visual inspection also allow to identify the side chain of a histidine residue involved in the hydrogen bonding with surrounding molecules and in that case the δ nitrogen of the histidine (HSB) is protonated residue.