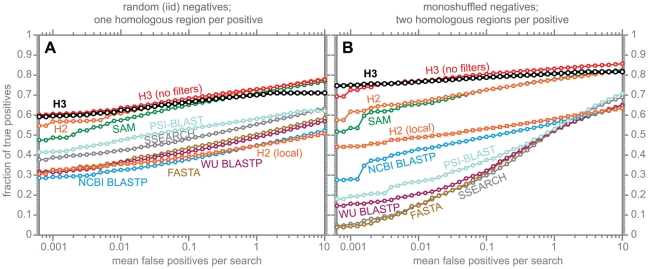
Figure 6. Benchmark of search sensitivity and specificity.
For different programs, searches are performed either by constructing a single profile from the query alignment (HMMER3, HMMER2, SAM, PSI-BLAST), or by using “family pairwise search” [41] in which each individual sequence is used as a query and the best E-value per target sequence is recorded (BLASTP, SSEARCH, FASTA). In each benchmark, true positive subsequences have been selected to be no more than 25% identical to any sequence in the query alignment (see Methods). Panel A shows results where nonhomologous sequence has been synthesized by a simple random model, and each true positive sequence contains a single embedded homologous subsequence (a total of 2,141 query multiple alignments, 11,547 true positive sequences, and 200,000 decoys). Panel B shows results where nonhomologous sequence is synthesized by shuffling randomly chosen subsequences from UniProt, and each true positive contains two embedded homologous subsequences (a total of 2,141 query alignments, 24,040 true positive sequences, and 200,000 decoys). The Y-axis is the fraction of true positives detected with an E-value better than the number of false positives per query specified on the X-axis.