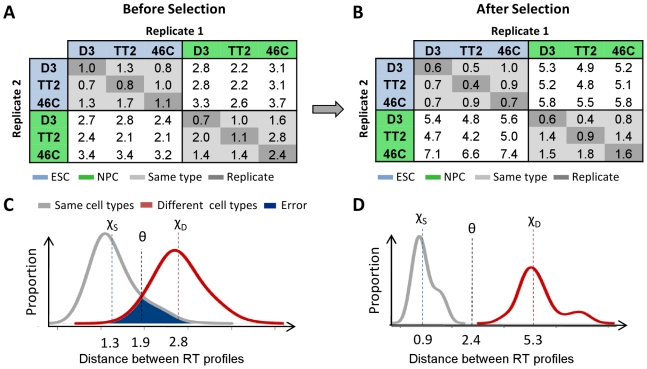
Figure 2. Monte Carlo optimization of fingerprinting regions.
A Monte Carlo algorithm is used to select regions with maximal differences in replication timing between cell types and minimal differences between replicates to obtain an optimized set of genomic regions for classification using the nearest-neighbor method. A,B. Selection of fingerprinting regions accentuates differences between cell types while diminishing those within equivalent cell types (light gray) and replicates (dark gray). C,D. To calculate confidence levels of predictions we use the distributions of distances within (grey) and between (red) cell types, shown here for 30 runs before and after selection. The error rate of prediction is represented by the blue shaded area shared by comparisons between similar or distinct cell types, with average distances of χS and χD respectively. The optimal classifier, θ, is estimated by minimizing the number of misclassified distances as in Figure 3 and Figure 4. Above this distance, datasets are predicted to originate from different cell types.