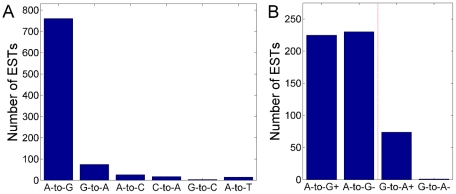
Figure 4. The number of ultra-editing events by mismatch type and strand.
(A) The number of ultra-edited ESTs of each mismatch type. The number of A-to-G ESTs is much larger than any other mismatch type, suggesting that the A-to-G clusters are due to A-to-I ultra-editing. Only six (out of 12) mismatch types are presented: ultra-editing of the complementary mismatches were mostly removed in the cleanup procedure. (B) The number of ultra-edited ESTs of type A-to-G and G-to-A, broken by the RNA strand. The (+) sign corresponds to the sequenced RNA being A or G; the (−) sign corresponds to T or C. For G-to-A, in all but one EST the (+) strand was edited, suggesting that many G-to-A ultra-edited ESTs may be due to a sequencing error. In this panel, we excluded 305 A-to-G edited ESTs arriving from a particular library (human liver regeneration after partial hepatectomy; see the main text), since in this library almost all ESTs (edited and non-edited) aligned to the sense strand. In the NCI-CGAP libraries, from which most of the G-to-A edited ESTs came, the sequenced RNA was biased towards the antisense strand, indicating that the difference between (+) and (−) demonstrated in the plot is not due to the experimental protocol.